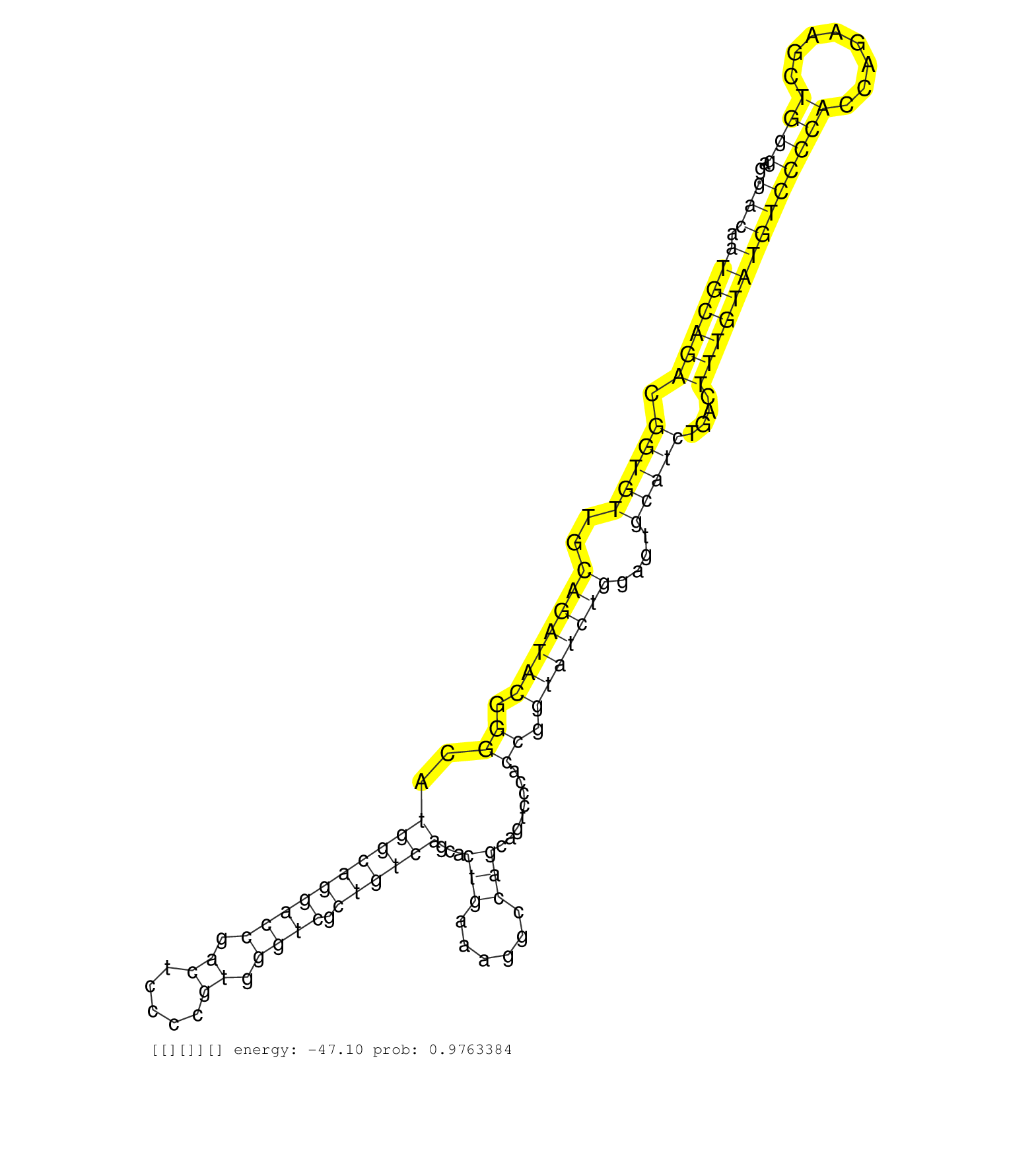
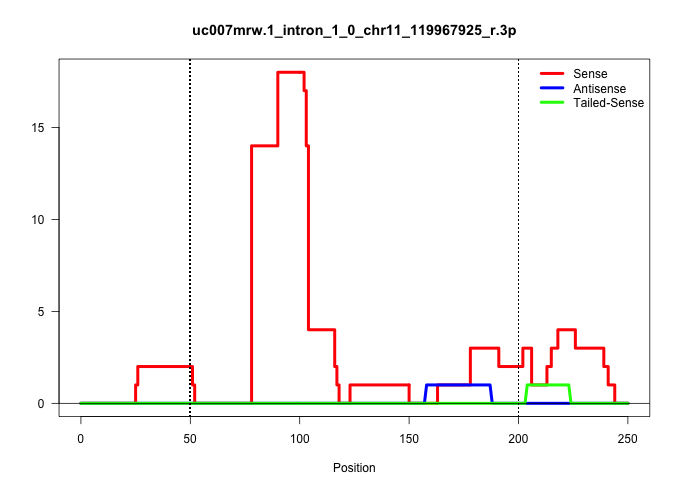
| Gene: 1810073N04Rik | ID: uc007mrw.1_intron_1_0_chr11_119967925_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(11) TESTES |
| CTGGTTAGGGCTTAGCCCTCTAGCCTTAACTCAGCGCCTCACTTCTGTGCCACTGGGACACTGGGACTGGGGTGACAATGCAGACGGTGTTGCAGATACGGGCATGGCAGGACCGACTCCCCGTGGGTCGCTGTCAGCACTGAAAGGCCAGCAGTCCCACCGGTATCTGGAGTGCATCTGACTTTGTATGTCCCCACCAGAAGCTGGGAGGGCAGAGATGCTGGACCATGCCGTGCTGCTACAGGTGATC .........................................................................(((.(((((((.(((((..(((((((.((..((((((((((.((.....)).)))).))))))...(((......)))........)).)))))))....)))))....))))))))))((((........)))).......................................... .........................................................................74......................................................................................................................................210...................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................TGCAGACGGTGTTGCAGATACGGGCA.................................................................................................................................................. | 26 | 1 | 10.00 | 10.00 | 2.00 | 1.00 | 2.00 | 3.00 | - | - | - | 1.00 | 1.00 | - | - |
| ..............................................................................TGCAGACGGTGTTGCAGATACGGGC................................................................................................................................................... | 25 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TGACTTTGTATGTCCCCACCAGAAGCTG............................................ | 28 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................................................TGCAGATACGGGCATGGCAGGACCGA...................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................GCTGGGAGGGCAGAGATGCTGGAC........................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...................................................................................................................................................................TATCTGGAGTGCATCTGACTTTGTATGT........................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..............................................................................TGCAGACGGTGTTGCAGATACGGG.................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .........................TTAACTCAGCGCCTCACTTCTGTGCC....................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .......................................................................................................................................................................................................................AGATGCTGGACCATGCCGTGCTGCTA......... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TGGGTCGCTGTCAGCACTGAAAGGCCA.................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................TGCAGATACGGGCATGGCAGGACCGAC..................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................TAACTCAGCGCCTCACTTCTGTGCCA...................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TGGGAGGGCAGAGATGtgct.......................... | 20 | tgct | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................................................................................TGCAGATACGGGCATGGCAGGACCGACT.................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................TGCTGGACCATGCCGTGCTGCTACAG...... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................AGAGATGCTGGACCATGCCGTGCTGC........... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| CTGGTTAGGGCTTAGCCCTCTAGCCTTAACTCAGCGCCTCACTTCTGTGCCACTGGGACACTGGGACTGGGGTGACAATGCAGACGGTGTTGCAGATACGGGCATGGCAGGACCGACTCCCCGTGGGTCGCTGTCAGCACTGAAAGGCCAGCAGTCCCACCGGTATCTGGAGTGCATCTGACTTTGTATGTCCCCACCAGAAGCTGGGAGGGCAGAGATGCTGGACCATGCCGTGCTGCTACAGGTGATC .........................................................................(((.(((((((.(((((..(((((((.((..((((((((((.((.....)).)))).))))))...(((......)))........)).)))))))....)))))....))))))))))((((........)))).......................................... .........................................................................74......................................................................................................................................210...................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................ACCGGTATCTGGAGTGCATCTGACTTTGTA.............................................................. | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................GGACCGACTCCCCGgag............................................................................................................................... | 17 | gag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |