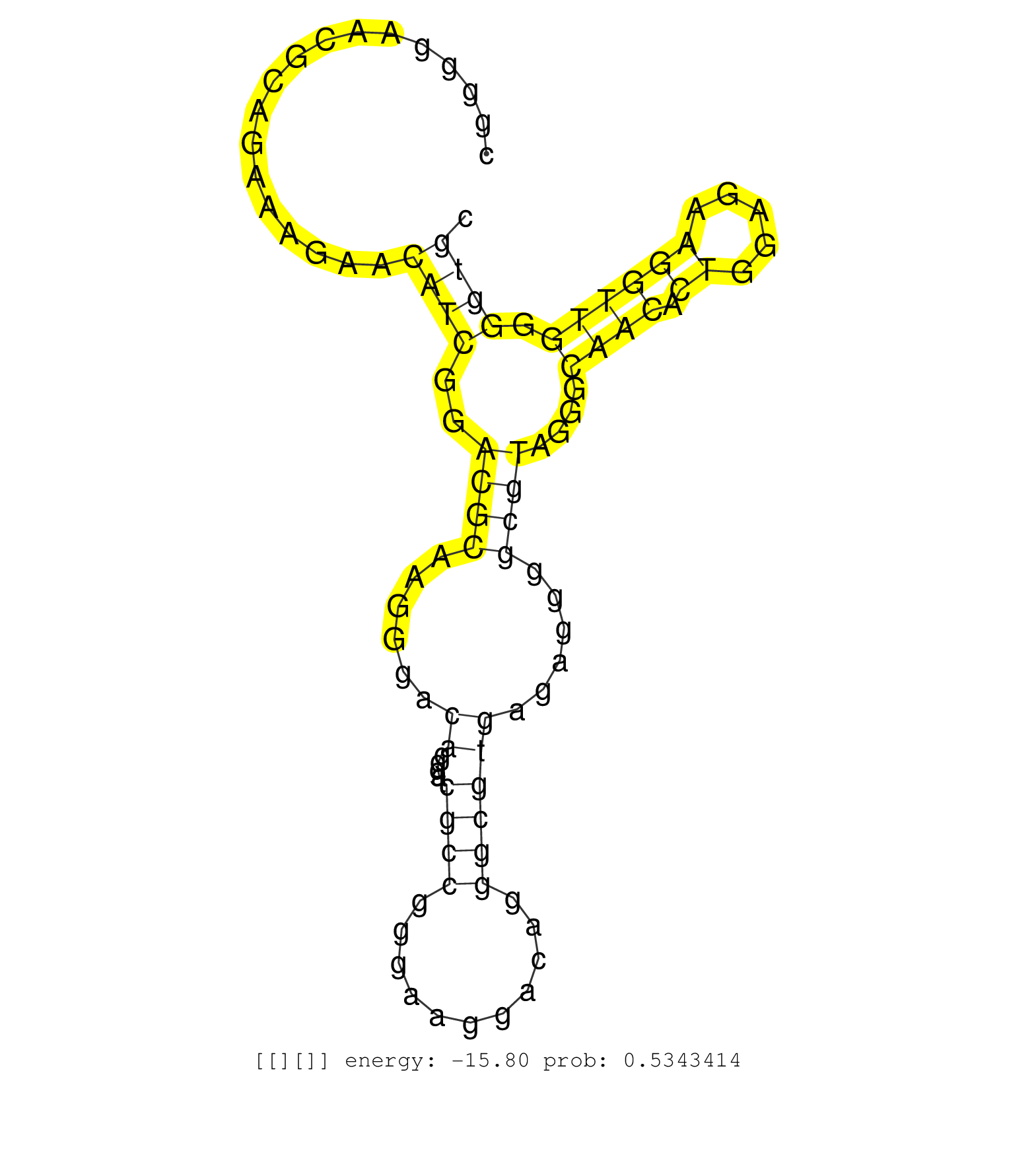

| Gene: mKIAA1453 | ID: uc007mot.1_intron_18_0_chr11_118149138_r.5p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(5) PIWI.ip |
(2) PIWI.mut |
(17) TESTES |
| GGGTGCGCGTTCTACCCGCGGGTTTTGGGGACTCCAGCGAAGGCTGACAGGTGCGTGCGTGGCCGGCGAGGGCACCGGGGAACGCAGAAAGAACATCGGACGCAAGGGACAGGGATCGCCGGGAAGGACAGGGCGTGAGAGGGGCGTAGGGCAACACTGGAGAAGGTTGGGGTGCGAGGGACTTGGAGGAACAAGGGATGCCAGGGACTCCGAGGAAGGGACAGGTGATGTGAGGGGTGCCTGTAAACCC .............................................................................................((((..((((......((.....((((...........))))))......))))....((((.((.....)))))).))))............................................................................ ...........................................................................76.................................................................................................175......................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................TGGGGACTCCAGCGAAGGCTGACAGGT...................................................................................................................................................................................................... | 27 | 1 | 4.00 | 4.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - |
| ................................................................................AACGCAGAAAGAACATCGGACGCAAGG............................................................................................................................................... | 27 | 1 | 4.00 | 4.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TAGGGCAACACTGGAGAAGGTTGGG............................................................................... | 25 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................AACGCAGAAAGAACATCGGACGCAAGGG.............................................................................................................................................. | 28 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................GCCGGGAAGGACAGGGCG................................................................................................................... | 18 | 2 | 2.00 | 2.00 | - | - | - | - | - | - | 0.50 | 1.50 | - | - | - | - | - | - | - | - | - |
| ................................................................................AACGCAGAAAGAACATCGGACGCAAGGGt............................................................................................................................................. | 29 | t | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................GAACATCGGACGCAAGGGACAGGGA....................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................TGGGGACTCCAGCGAAGGCTGACAGG....................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................ACCGGGGAACGCAGAAAGAACA........................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................GAACAAGGGATGCCAGGGACTCCGAGGA.................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................GACGCAAGGGACAGGGATCGCCGGG............................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................TGGGGACTCCAGCGAAGGCTGACAGGTG..................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................GAACGCAGAAAGAACATCGGACGCAAGG............................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................................AAGGCTGACAGGTGCGTGCGTGGCCGGC....................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................GGGGACTCCAGCGAAGGCTGACAGGTG..................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................................................................................................................................................GGACTTGGAGGAACAAGGG..................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................CAGAAAGAACATCGGACGCAAGGGAC............................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................GGGACTCCAGCGAAGGCTGACAGGTG..................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................AACGCAGAAAGAACATCGGACGCAAGGGA............................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................TCCGAGGAAGGGACAGGTGATGTGAGG............... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................GGACAGGGATCGCCGGGAAGGACAGG...................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................................CACCGGGGAACGCAGAAAGAACATCGG....................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............CCGCGGGTTTTGGGGACT......................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ......................TTTTGGGGACTCCAGCGAAGGCTGAC.......................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .......................TTTGGGGACTCCAGCGAAGGCTGACAG........................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........GTTCTACCCGCGGGTTTTGGGGACT......................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................TGGAGAAGGTTGGGGTGCGAGGGAC.................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................GGACAGGGATCGCCGGGA.............................................................................................................................. | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - |
| GGGTGCGCGTTCTACCCGCGGGTTTTGGGGACTCCAGCGAAGGCTGACAGGTGCGTGCGTGGCCGGCGAGGGCACCGGGGAACGCAGAAAGAACATCGGACGCAAGGGACAGGGATCGCCGGGAAGGACAGGGCGTGAGAGGGGCGTAGGGCAACACTGGAGAAGGTTGGGGTGCGAGGGACTTGGAGGAACAAGGGATGCCAGGGACTCCGAGGAAGGGACAGGTGATGTGAGGGGTGCCTGTAAACCC .............................................................................................((((..((((......((.....((((...........))))))......))))....((((.((.....)))))).))))............................................................................ ...........................................................................76.................................................................................................175......................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|