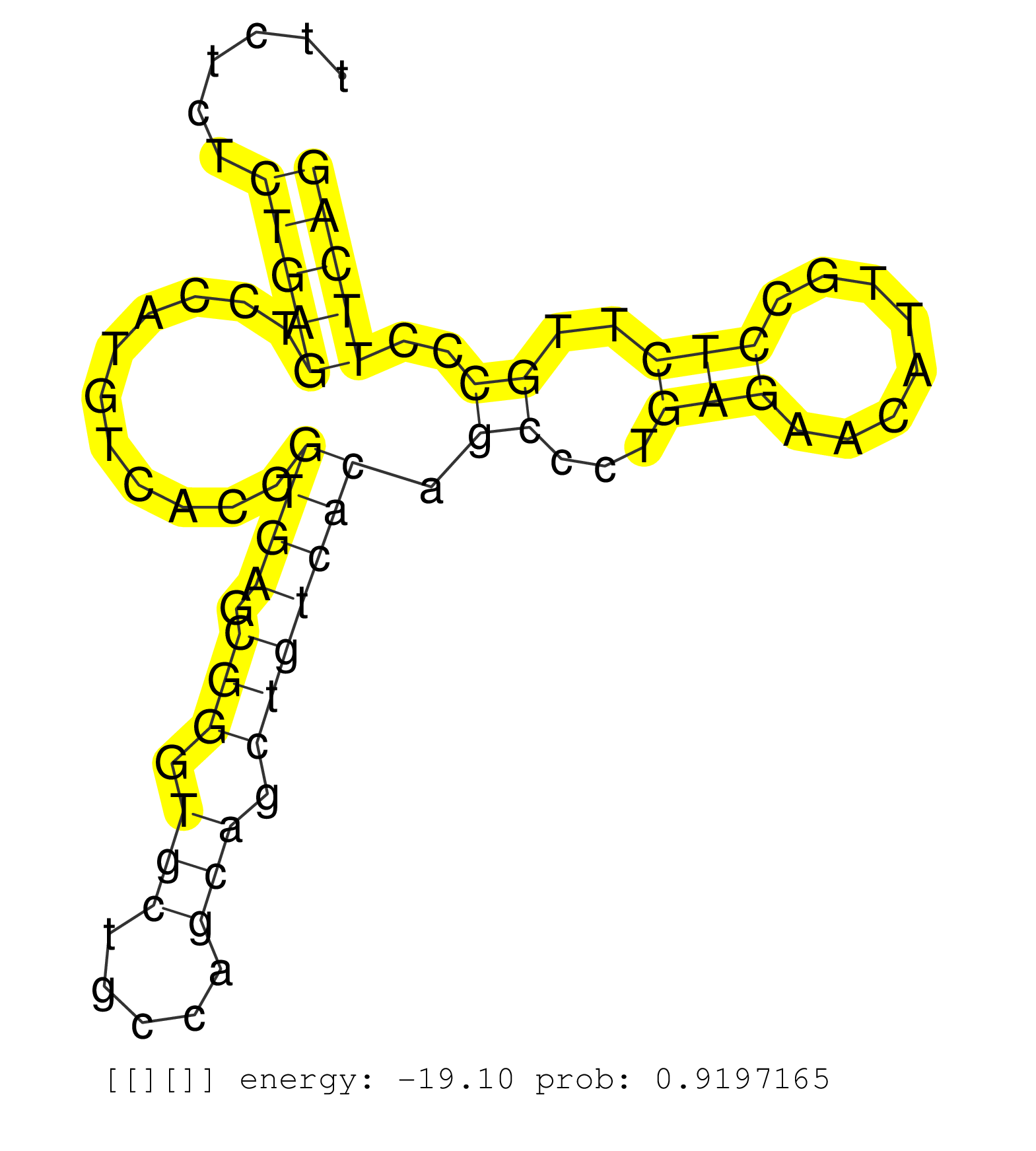

| Gene: mKIAA0585 | ID: uc007mmh.1_intron_1_0_chr11_116700443_r.3p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(7) PIWI.ip |
(1) PIWI.mut |
(24) TESTES |
| ACAGGCGCGTGGCTCTAGAATGTTGTTTTCCTTCAGCGAGTGTGGACAGCCTGTGTGGTCTGTCCCAGCCCAGTGGTGCCACCTGGAGCCACTTCCAGGTGCTGTAAGCAATGGAGGCCTTCTCTCTGAGTCCATGTCACCGTGAGCGGGTGCTGCCAGCAGCTGTCACAGCCCTGAGAACATTGCCTCTTGCCCTTCAGGATCTTGAAACAGGAGCACCCTGAGCTGGCAGTCCTGGCCGACGCAGTTG .............................................................................................................................(((((...........((((.(((.(((.....))).))))))).((...(((........)))..))..))))).................................................. .......................................................................................................................120.............................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............TAGAATGTTGTTTTCCTTCAGCGAGT................................................................................................................................................................................................................. | 26 | 1 | 5.00 | 5.00 | - | - | 2.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TGAAACAGGAGCACCCTGAGCTGGCAG.................. | 27 | 1 | 3.00 | 3.00 | - | - | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................TTTCCTTCAGCGAGTGTGGACAGCCTG..................................................................................................................................................................................................... | 27 | 1 | 3.00 | 3.00 | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TGAAACAGGAGCACCCTGAGCTGGCAGTC................ | 29 | 1 | 3.00 | 3.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................TGCCTCTTGCCCTTCAGGATCTTGAAAC....................................... | 28 | 1 | 3.00 | 3.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .............................................................................................................................................................................................................TGAAACAGGAGCACCCTGAGCTGGC.................... | 25 | 1 | 3.00 | 3.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................TTGCCCTTCAGGATCTTGAAACAGGAGC................................. | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - |
| .............................................................................................................................................................................................................TGAAACAGGAGCACCCTGAGCTGGCA................... | 26 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............TAGAATGTTGTTTTCCTTCAGCGAGTGT............................................................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................TCTGAGTCCATGTCACCGTGAGCGGGT................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TCTTGAAACAGGAGCACCCTGAGCTGGC.................... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 |
| ..............................................................................................................................TGAGTCCATGTCACCGTGAGCGGGTGC................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................TCTCTGAGTCCATGTCACCGTGAGCGGGT................................................................................................... | 29 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................TCTCTGAGTCCATGTCACCGTGAGCGG..................................................................................................... | 27 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGAGAACATTGCCTCTTGCCCTTCAG.................................................. | 26 | 1 | 2.00 | 2.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................TGGTCTGTCCCAGCCCAGTGGTGCCACC....................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGTGTGGTCTGTCCCAGCCCAGTGGTG............................................................................................................................................................................ | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................TCAGGATCTTGAAACAGGAGCACC.............................. | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................CCAGGTGCTGTAAGCtt........................................................................................................................................... | 17 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................GGAGCACCCTGAGCTGGCAGTC................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................AAACAGGAGCACCCTGAGCTGGCAGTCCaat............ | 31 | aat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................ACAGGAGCACCCTGAGCTGGCAGgcct.............. | 27 | gcct | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................TGCCCTTCAGGATCTTGAAACAGGAGC................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................TTGCCTCTTGCCCTTCAGGAca.............................................. | 22 | ca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................TGTCACAGCCCTGAGAACATTGCCTCT............................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TGAAACAGGAGCACCCTGAGCTGGCAGgcc............... | 30 | gcc | 1.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TCTTGAAACAGGAGCACCCTGAGCTGG..................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................AAACAGGAGCACCCTGAGCTGGCAGTC................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................TGTCACAGCCCTGAGAACATTGCCTCTTGC......................................................... | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................GAACATTGCCTCTTGCCCTTCAGGATC.............................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................TTGCCCTTCAGGATCTTGAAACAGGAG.................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................CTGAGCTGGCAGTCCTGGCCGACG...... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...............TAGAATGTTGTTTTCCTTCAGCGtgtg................................................................................................................................................................................................................ | 27 | tgtg | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................TGTCACAGCCCTGAGAACATTGCCTCTT........................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................TTCCAGGTGCTGTAAGCAATGGAGGCCT.................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................TCTGAGTCCATGTCACCGTGAGCGGGTGCTGC.............................................................................................. | 32 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............TAGAATGTTGTTTTCCTTCAGCGAGTG................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................GAACATTGCCTCTTGCCCTTCAGGAT............................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................CTTGAAACAGGAGCACCCTGAGCTGGCAG.................. | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................CAGGATCTTGAAACAGGAGCACC.............................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .............................................................................................TCCAGGTGCTGTAAGCAATGGAGGCCT.................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TGAGTCCATGTCACCGTGAGCGGGTt.................................................................................................. | 26 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGGATCTTGAAACAGGAGCACCCTGAGC........................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................TGAGCTGGCAGTCCTGGC........... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ......................................................................................................TGTAAGCAATGGAGGCCTTCTCTCTGAa........................................................................................................................ | 28 | a | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................GAACATTGCCTCTTGCCCTTCAGGATCT............................................. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................CTCTGAGTCCATGTCACCGTGAGCGG..................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TCCAGGTGCTGTAAGCAATGGAGGCCTTCTC.............................................................................................................................. | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TGAAACAGGAGCACCCTGAGCTGGCAGT................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................TGCCAGCAGCTGTCACAGCCCTGAGA....................................................................... | 26 | 2 | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................TGCCAGCAGCTGTCACAGCCCTGAG........................................................................ | 25 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TGAGCGGGTGCTGCCAGCAGCTGTCAC................................................................................. | 27 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................TAAGCAATGGAGGCCTTCTCTCTGAGTC...................................................................................................................... | 28 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................TGTGGTCTGTCCCAGC..................................................................................................................................................................................... | 16 | 4 | 0.25 | 0.25 | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ACAGGCGCGTGGCTCTAGAATGTTGTTTTCCTTCAGCGAGTGTGGACAGCCTGTGTGGTCTGTCCCAGCCCAGTGGTGCCACCTGGAGCCACTTCCAGGTGCTGTAAGCAATGGAGGCCTTCTCTCTGAGTCCATGTCACCGTGAGCGGGTGCTGCCAGCAGCTGTCACAGCCCTGAGAACATTGCCTCTTGCCCTTCAGGATCTTGAAACAGGAGCACCCTGAGCTGGCAGTCCTGGCCGACGCAGTTG .............................................................................................................................(((((...........((((.(((.(((.....))).))))))).((...(((........)))..))..))))).................................................. .......................................................................................................................120.............................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|