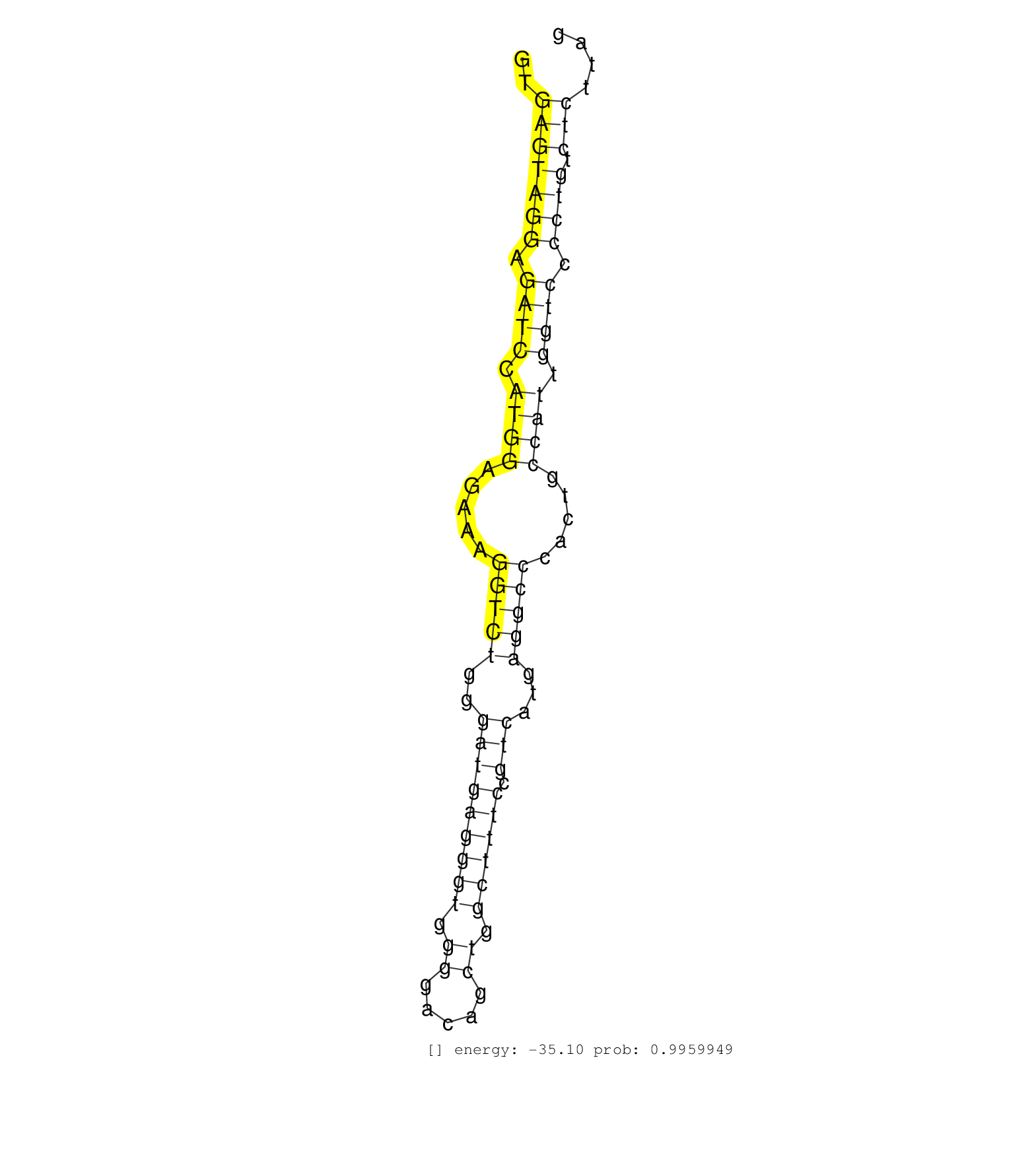
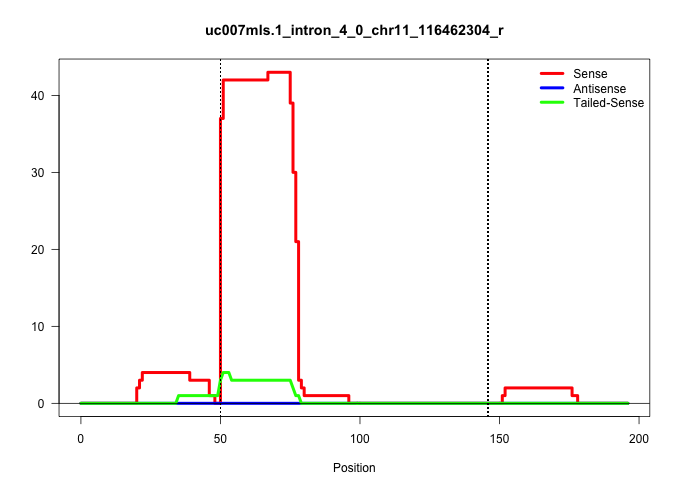
| Gene: Rhbdf2 | ID: uc007mls.1_intron_4_0_chr11_116462304_r | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(5) PIWI.ip |
(2) PIWI.mut |
(13) TESTES |
| CTGCCTCCAGTGGGGCCCACATCTGGCCTGATGACATTACCAAGTGGCCGGTGAGTAGGAGATCCATGGAGAAAGGTCTGGGATGAGGGTGGGGACAGCTGGCTTTCCGTCATGAGGCCCACTGCCATTGGTCCCCTGTCTCTTAGATCTGCACAGAGCAGGCTCAGAGCAACCACACGGGCTTGTTGCACATAGA ....................................................(((((((.((((.((((.....(((((..(((((((((.((.....)).)))))).)))...))))).....)))).)))).)))).)))...................................................... ..................................................51.............................................................................................146................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGAGTAGGAGATCCATGGAGAAAGGTC...................................................................................................................... | 28 | 1 | 16.00 | 16.00 | 8.00 | 4.00 | 1.00 | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - |
| ..................................................GTGAGTAGGAGATCCATGGAGAAAGG........................................................................................................................ | 26 | 1 | 8.00 | 8.00 | 3.00 | - | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - |
| ..................................................GTGAGTAGGAGATCCATGGAGAAAGGT....................................................................................................................... | 27 | 1 | 8.00 | 8.00 | 1.00 | 1.00 | 3.00 | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - |
| ..................................................GTGAGTAGGAGATCCATGGAGAAAG......................................................................................................................... | 25 | 1 | 4.00 | 4.00 | 1.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...................................................TGAGTAGGAGATCCATGGAGAAAGGTC...................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................................................................................................................................ACAGAGCAGGCTCAGAGCAACCAC.................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................TGAGTAGGAGATCCATGGAGAAAGGTCT..................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...................................................TGAGTAGGAGATCCATGGAGAAAGGT....................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................ATCTGGCCTGATGACATTA............................................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................GGAGAAAGGTCTGGGATGAGGGTGGGGAC.................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTAGGAGATCCATGGAGAAAGGc....................................................................................................................... | 27 | c | 1.00 | 8.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGTAGGAGATCCATGGAGAAAGGTaa..................................................................................................................... | 28 | aa | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................CTGGCCTGATGACATTACCAAGTG...................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................ATTACCAAGTGGCCGatct.............................................................................................................................................. | 19 | atct | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTAGGAGATCCATGGAGAAAGt........................................................................................................................ | 26 | t | 1.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................ATCTGGCCTGATGACATTACCAAGTG...................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................CACAGAGCAGGCTCAGAGCAACCACAC.................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .....................TCTGGCCTGATGACATTACCAAGTGGC.................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGTAGGAGATCCATGGAGAAAGG........................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTAGGAGATCCATGGAGAAAGGTCTG.................................................................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| CTGCCTCCAGTGGGGCCCACATCTGGCCTGATGACATTACCAAGTGGCCGGTGAGTAGGAGATCCATGGAGAAAGGTCTGGGATGAGGGTGGGGACAGCTGGCTTTCCGTCATGAGGCCCACTGCCATTGGTCCCCTGTCTCTTAGATCTGCACAGAGCAGGCTCAGAGCAACCACACGGGCTTGTTGCACATAGA ....................................................(((((((.((((.((((.....(((((..(((((((((.((.....)).)))))).)))...))))).....)))).)))).)))).)))...................................................... ..................................................51.............................................................................................146................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|