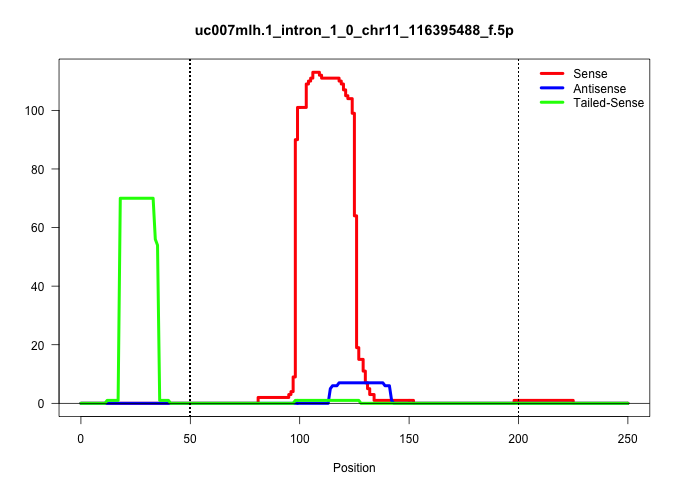
| Gene: Sphk1 | ID: uc007mlh.1_intron_1_0_chr11_116395488_f.5p | SPECIES: mm9 |
 |
 |
|
(6) OTHER.mut |
(2) OVARY |
(5) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(27) TESTES |
| ACCGGTGGCCCCTTGTCAGCGGGAGCCCCGGGACCTGGCTATGGAACCAGGTTTGTGGGGTACCTGTCTGGGAAAGGCTAATGAGATCTCTTCCCCCAAGAAATGATGGACAGCAGAAAGAAGCCTAGGGTCACTCGAAAGAAACATTTTGTCCCGCGAGGCCAGACATCGTCTGTCCCGGGGCCAAGGAAAGAGGTATTGCGGTGTCCCACTCACTCCCAGACAACCTGTGGGAGGCCACTTCCTCTGG ..........................................................................................................................((((....((..((....))......(((.(((.((.((.(((((...))))).))))))).))).))...))))..................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR037898(GSM510434) ovary_rep3. (ovary) | SRR037897(GSM510433) ovary_rep2. (ovary) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR037903(GSM510439) testes_rep4. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................GCGGGAGCCCCGGGgaga...................................................................................................................................................................................................................... | 18 | gaga | 58.00 | 0.00 | - | - | - | - | 13.00 | 8.00 | 10.00 | - | - | - | - | - | - | - | 5.00 | 4.00 | 2.00 | - | 3.00 | 3.00 | 3.00 | - | 3.00 | - | 1.00 | 1.00 | 2.00 | - | - |
| ..................................................................................................AGAAATGATGGACAGCAGAAAGAAGCCT............................................................................................................................ | 28 | 1 | 38.00 | 38.00 | 8.00 | 8.00 | 6.00 | 9.00 | - | - | - | - | 1.00 | 1.00 | 1.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................................................................AGAAATGATGGACAGCAGAAAGAAGCC............................................................................................................................. | 27 | 1 | 33.00 | 33.00 | 6.00 | 6.00 | 5.00 | 7.00 | - | - | - | 1.00 | - | - | - | 3.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | 1.00 | - |
| ..................GCGGGAGCCCCGGGga........................................................................................................................................................................................................................ | 16 | ga | 14.00 | 0.00 | 1.00 | - | 1.00 | - | - | - | - | 6.00 | 4.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................GAAATGATGGACAGCAGAAAGAAGCCT............................................................................................................................ | 27 | 1 | 7.00 | 7.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................AGAAATGATGGACAGCAGAAAGAAGC.............................................................................................................................. | 26 | 1 | 4.00 | 4.00 | - | - | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TGATGGACAGCAGAAAGAAGCCTAGGG........................................................................................................................ | 27 | 1 | 4.00 | 4.00 | - | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................AGAAATGATGGACAGCAGAAAGAAGCCTA........................................................................................................................... | 29 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................GCGGGAGCCCCGGGtaga...................................................................................................................................................................................................................... | 18 | taga | 3.00 | 0.00 | - | - | - | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................AAGAAATGATGGACAGCAGAAAGAAGCC............................................................................................................................. | 28 | 1 | 3.00 | 3.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TGGACAGCAGAAAGAAGCCTAGGGTCAC.................................................................................................................... | 28 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......................................................................................................TGATGGACAGCAGAAAGAAGCCTAGG......................................................................................................................... | 26 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................AGAAATGATGGACAGCAGAAAGAAGCCTAGG......................................................................................................................... | 31 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................GAAATGATGGACAGCAGAAAGAAGCC............................................................................................................................. | 26 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................GCGGGAGCCCCGGGgatc...................................................................................................................................................................................................................... | 18 | gatc | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................GCGGGAGCCCCGGGgag....................................................................................................................................................................................................................... | 17 | gag | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................AAGAAATGATGGACAGCAGAAAGAAGC.............................................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................AGAAATGATGGACAGCAGAA.................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................GCGGGAGCCCCGGGgcga...................................................................................................................................................................................................................... | 18 | gcga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ......................................................................................................................................................................................................TTGCGGTGTCCCACTCACTCCCAGACA......................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ............TTGTCAGCGGGAGCCCCGGGACCTGGCaa................................................................................................................................................................................................................. | 29 | aa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................TGAGATCTCTTCCCCCAAGAAATGATGGA............................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................AGAAATGATGGACAGCAGAAAGAA................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TGATGGACAGCAGAAAGAAGCCTAGGGT....................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................GAAATGATGGACAGCAGAAAGAAGCCTA........................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................GCGGGAGCCCCGGGgata...................................................................................................................................................................................................................... | 18 | gata | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................AAGAAATGATGGACAGCAGAAAG.................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TGATGGACAGCAGAAAGAAGCCTAGGGTC...................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................GAAATGATGGACAGCAGAAAGA................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................CAGCGGGAGCCCCGGGga........................................................................................................................................................................................................................ | 18 | ga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................AGAAATGATGGACAGCAGAAAGAAGCCTAa.......................................................................................................................... | 30 | a | 1.00 | 3.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................ATGGACAGCAGAAAGAAGCCTAGGGTC...................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCAAGAAATGATGGACAGCAGAAAG.................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................TGAGATCTCTTCCCCCAAGAAATGATGG............................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................CAAGAAATGATGGACAGCAGAAAGA................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................TAGGGTCACTCGAAAGAAACATTTTGT.................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................AGAAATGATGGACAGCAGAAA................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................GATGGACAGCAGAAAGAAGCCTAGGGT....................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ACCGGTGGCCCCTTGTCAGCGGGAGCCCCGGGACCTGGCTATGGAACCAGGTTTGTGGGGTACCTGTCTGGGAAAGGCTAATGAGATCTCTTCCCCCAAGAAATGATGGACAGCAGAAAGAAGCCTAGGGTCACTCGAAAGAAACATTTTGTCCCGCGAGGCCAGACATCGTCTGTCCCGGGGCCAAGGAAAGAGGTATTGCGGTGTCCCACTCACTCCCAGACAACCTGTGGGAGGCCACTTCCTCTGG ..........................................................................................................................((((....((..((....))......(((.(((.((.((.(((((...))))).))))))).))).))...))))..................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR037898(GSM510434) ovary_rep3. (ovary) | SRR037897(GSM510433) ovary_rep2. (ovary) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR037903(GSM510439) testes_rep4. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................AGAAAGAAGCCTAGGGTCACTCGAAAGA............................................................................................................ | 28 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - |
| ..................................................................................................................AAGAAGCCTAGGGTCACTCcct.................................................................................................................. | 22 | cct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ....................................................................................................................AAGAAGCCTAGGGTCACTa................................................................................................................... | 19 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................GAAAGAAGCCTAGGGTCACTCGAAAGA............................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ............................................................................................................................TAGGGTCACTCGAAAGAAAa.......................................................................................................... | 20 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................AGAAATGATGGACActac.......................................................................................................................................... | 18 | ctac | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................AGAAGCCTAGGGTCACTCGAAAGAA........................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................AGGCCACTTCCTCTGgt. | 17 | gt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................AGAAAGAAGCCTAGGGTCACTCGAA............................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |