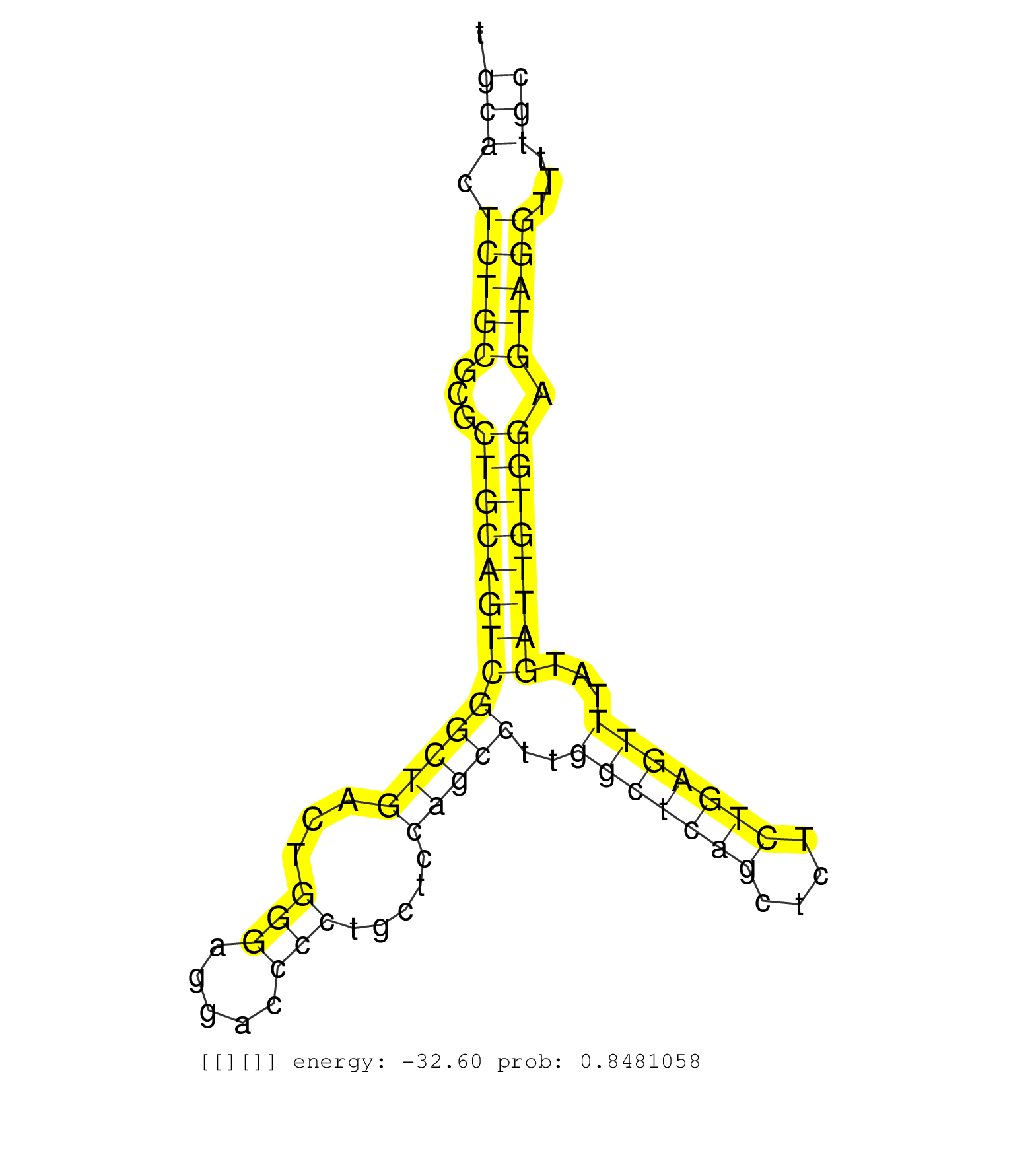

| Gene: Nup85 | ID: uc007mhx.1_intron_6_0_chr11_115431435_f.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(1) OVARY |
(4) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(14) TESTES |
| ATGTCCTGGGCGGTGACAACCCAAGCAAGCATGAAAACTTCTGGGACCTGGTGAGTGTGCGCGGACGGCAGCCGCCCGCCCTGTGCACTCTGCGCGCTGCAGTCGGCTGACTGGGAGGACCCCTGCTCCAGCCTTGGCTCAGCTCTCTGAGTTTATGATTGTGGAGTAGGTTTTGCATCTTTCCTTCTCTGCTCCAGAGCAGTTTAAAACTAATTTAGCACGGGGAGCGGTGCTTGGTAAACTTGCCGTC ....................................................................................(((.(((((...(((((((((((((...(((.....))).....)))))..(((((((....)))))))...)))))))).)))))...))).......................................................................... ...................................................................................84..........................................................................................176........................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................TCTGAGTTTATGATTGTGGAGTAGGTT.............................................................................. | 27 | 1 | 10.00 | 10.00 | 6.00 | 2.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...................................................................................................................................................TGAGTTTATGATTGTGGAGTAGGTT.............................................................................. | 25 | 1 | 4.00 | 4.00 | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - |
| ...............................TGAAAACTTCTGGGACCTGGTGAGT.................................................................................................................................................................................................. | 25 | 1 | 3.00 | 3.00 | - | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................TGAAAACTTCTGGGACCTGGTGActg................................................................................................................................................................................................. | 26 | ctg | 2.00 | 0.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TGAGTTTATGATTGTGGAGTAGGTTT............................................................................. | 26 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................TCTGCGCGCTGCAGTCGGCTGACTGGt....................................................................................................................................... | 27 | t | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................GTGGAGTAGGTTTccgg......................................................................... | 17 | ccgg | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................TGCACTCTGCGCGCTGCAGTCGGCTGt............................................................................................................................................ | 27 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......................AGCAAGCATGAAAACTTCTGGGACCTGt....................................................................................................................................................................................................... | 28 | t | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................GCAAGCATGAAAACTTCTGGGACCTGG....................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................CAAGCATGAAAACTTCTGGGACCTGGT...................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................GCAAGCATGAAAACTTCTGGGACCTGGc...................................................................................................................................................................................................... | 28 | c | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................AACTTCTGGGACCTGGTGAct.................................................................................................................................................................................................. | 21 | ct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................................................................TCTGCGCGCTGCAGTCGGCTGACTGc........................................................................................................................................ | 26 | c | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................AGCAAGCATGAAAACTTCTGGGACCT......................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................CCTGTGCACTCTGCGCGCTGCAGTCGGC............................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TCTGAGTTTATGATTGTGGAGTAGGT............................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................ATCTTTCCTTCTCTGCTCCAGAGCAGT............................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...........................................................................................................................................................TGATTGTGGAGTAGGTTTTGCATCTT..................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................TGCACTCTGCGCGCTGCAGTCGGCT.............................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGTGTGCGCGGACGGCAGCCGCCCG............................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................TGAAAACTTCTGGGACCTGGTGAca.................................................................................................................................................................................................. | 25 | ca | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................TGTGCACTCTGCGCGCTGCAGTC.................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................................TGTGCACTCTGCGCGCTGCAGTCGGC............................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................CTCTGCGCGCTGCAGTCGGCTGACTGG........................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................TATGATTGTGGAGTAGGTTTTGCATC....................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................TGTGCACTCTGCGCGCTGCAGTCGGCT.............................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................GCAAGCATGAAAACTTCTGGGACCTG........................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................AAGCATGAAAACTTCTGGGACCTGGTG..................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......................................................................................................................................................GTTTATGATTGTGGAGTAGGTTTTt........................................................................... | 25 | t | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................TGAAAACTTCTGGGACCTGGTGAc................................................................................................................................................................................................... | 24 | c | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ............................................................................................................................................................................TTGCATCTTTCCTTCTCTGCTCCAGAGt.................................................. | 28 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ATGTCCTGGGCGGTGACAACCCAAGCAAGCATGAAAACTTCTGGGACCTGGTGAGTGTGCGCGGACGGCAGCCGCCCGCCCTGTGCACTCTGCGCGCTGCAGTCGGCTGACTGGGAGGACCCCTGCTCCAGCCTTGGCTCAGCTCTCTGAGTTTATGATTGTGGAGTAGGTTTTGCATCTTTCCTTCTCTGCTCCAGAGCAGTTTAAAACTAATTTAGCACGGGGAGCGGTGCTTGGTAAACTTGCCGTC ....................................................................................(((.(((((...(((((((((((((...(((.....))).....)))))..(((((((....)))))))...)))))))).)))))...))).......................................................................... ...................................................................................84..........................................................................................176........................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|