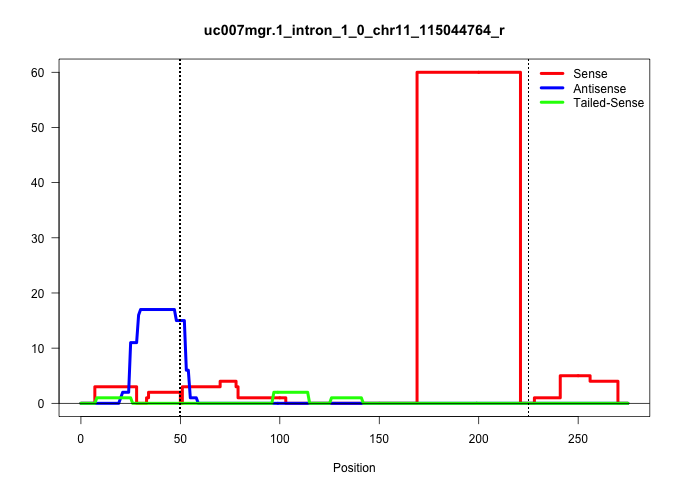
| Gene: Nat9 | ID: uc007mgr.1_intron_1_0_chr11_115044764_r | SPECIES: mm9 |
 |
|
(2) OTHER.mut |
(1) PIWI.mut |
(12) TESTES |
| CAGGCGGCAGGGACTTGGCACTGAGGCGTCTCTCCTGATCATGTCCTACGGTAAGACAGACAGAACTGACCACATGGGTAGAGGTGGGCAGAGCTCAGCTGAACCTCTGGGGGTGGTGGGGGAAACAGCAGATGACAAGCCCCCAACTGCAGACTGGGACCAGAGGCTAGAAGAGTAGAGCTTTTACTGTGTTCTTCTCCCCTCCCCGCCCCCCTGTAACTGGAGGTGTGACGAAGTTAGGTCTGACCAAGTTTGAGGCCAAAATCGGGCAAGAA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesWT4() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | mjTestesWT3() Testes Data. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................GAAGAGTAGAGCTTTTACTGTGTTCTTCTCCCCTCCCCGCCCCCCTGTAACT...................................................... | 52 | 1 | 60.00 | 60.00 | 60.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................................TCTGACCAAGTTTGAGGCCAAAATCGGGC..... | 29 | 1 | 4.00 | 4.00 | - | 4.00 | - | - | - | - | - | - | - | - | - | - |
| .......CAGGGACTTGGCACTGAGGCG....................................................................................................................................................................................................................................................... | 21 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................GCTGAACCTCTGGGGctt................................................................................................................................................................ | 18 | ctt | 2.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - |
| ...................................................TAAGACAGACAGAACTGACCACATGGGT.................................................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - |
| ..............................................................................................................................AGCAGATGACAAGCga..................................................................................................................................... | 16 | ga | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................CCTGATCATGTCCTACG................................................................................................................................................................................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................TAAGACAGACAGAACTGACCACATGGG..................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ........AGGGACTTGGCACTGgag......................................................................................................................................................................................................................................................... | 18 | gag | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................CACATGGGTAGAGGTGGGCAGAGCTCAGCTGAA............................................................................................................................................................................ | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................CTGATCATGTCCTACG................................................................................................................................................................................................................................. | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................TGACGAAGTTAGGTCTGACCAAGTTTGA................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| CAGGCGGCAGGGACTTGGCACTGAGGCGTCTCTCCTGATCATGTCCTACGGTAAGACAGACAGAACTGACCACATGGGTAGAGGTGGGCAGAGCTCAGCTGAACCTCTGGGGGTGGTGGGGGAAACAGCAGATGACAAGCCCCCAACTGCAGACTGGGACCAGAGGCTAGAAGAGTAGAGCTTTTACTGTGTTCTTCTCCCCTCCCCGCCCCCCTGTAACTGGAGGTGTGACGAAGTTAGGTCTGACCAAGTTTGAGGCCAAAATCGGGCAAGAA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesWT4() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | mjTestesWT3() Testes Data. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................GCGTCTCTCCTGATCATGTCCTACGGTA.............................................................................................................................................................................................................................. | 28 | 1 | 9.00 | 9.00 | - | - | 7.00 | 1.00 | - | - | - | - | - | - | - | 1.00 |
| .............................CTCTCCTGATCATGTCCTACGGTAAG............................................................................................................................................................................................................................ | 26 | 1 | 5.00 | 5.00 | - | 5.00 | - | - | - | - | - | - | - | - | - | - |
| ....................CTGAGGCGTCTCTCCTGATCATGTCCTA................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .....................TGAGGCGTCTCTCCTGATCATGTCCTA................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................TCTCCTGATCATGTCCTACGGTAAGACAG........................................................................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |