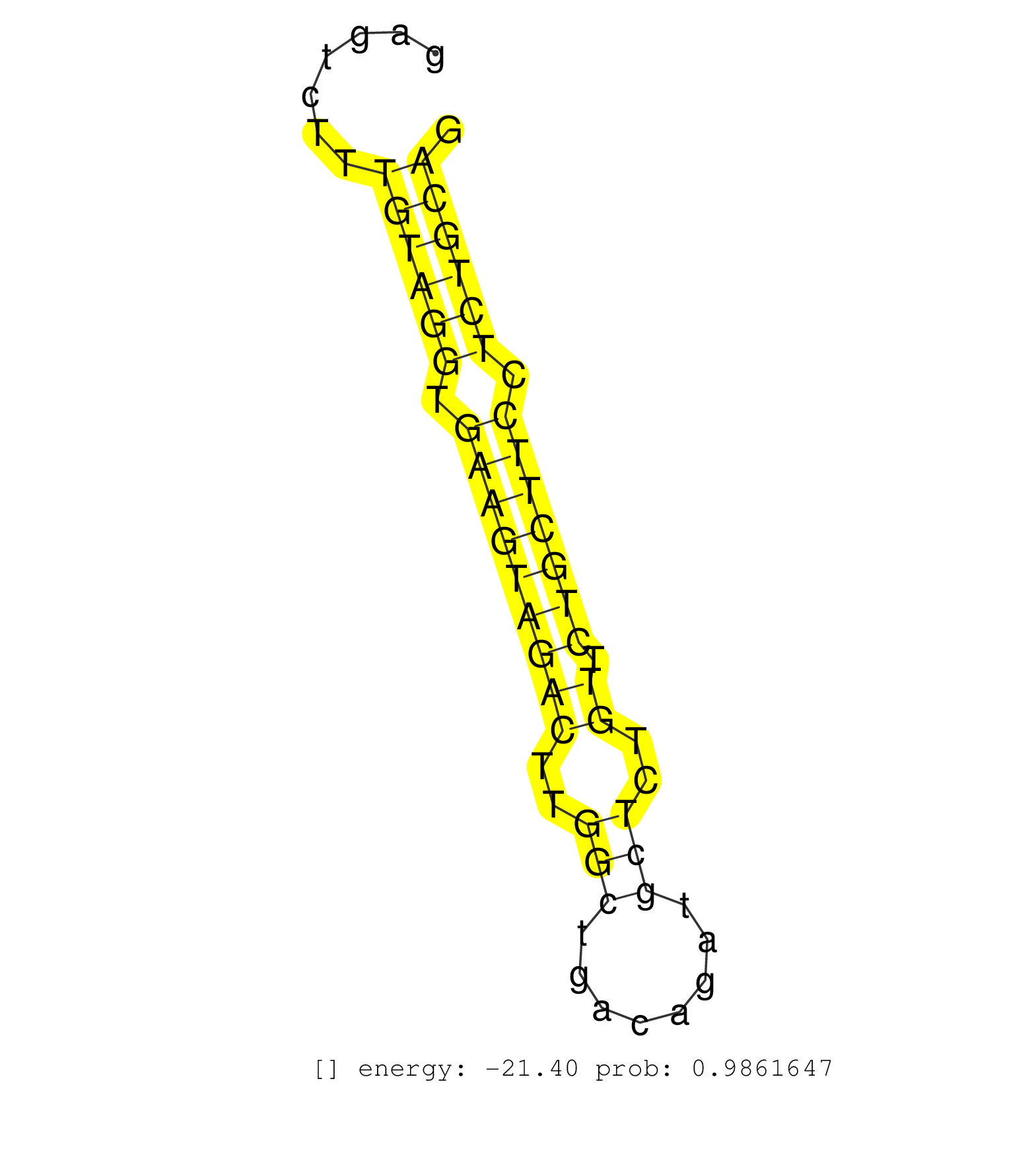

| Gene: Helz | ID: uc007mau.1_intron_19_0_chr11_107497626_f.3p | SPECIES: mm9 |
 |
 |
|
(6) OTHER.mut |
(1) OVARY |
(1) PIWI.ip |
(1) PIWI.mut |
(20) TESTES |
| AAAATCAATGCATTTAACAATGGTGCCATAGGGACCTTCTCTGTTATCACTGTCAGTGTTGTTAGGAAATCATTGGCGTCACAGGTATTCAGTGTTGGGACAAAATTAATTGTGGGCATAAATTAGTGTTGGAATTGGATGGAGTCTTTGTAGGTGAAGTAGACTTGGCTGACAGATGCTCTGTTCTGCTTCCTCTGCAGCTACACCTCTGAGCTTTTCTATGAGGGGAAGCTGATGGCCAGTGGGAAGC ....................................................................................................................................................((((((.(((((((((..(((........)))..)).))))))).))))))................................................... .............................................................................................................................................142.......................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesWT1() Testes Data. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................TCTGTTCTGCTTCCTCTGCAG.................................................. | 21 | 1 | 27.00 | 27.00 | - | 14.00 | 5.00 | 1.00 | - | 1.00 | 4.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TTTGTAGGTGAAGTAGACTTGGC................................................................................. | 23 | 1 | 13.00 | 13.00 | - | 9.00 | - | - | 1.00 | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................TGTAGGTGAAGTAGACTTGGC................................................................................. | 21 | 1 | 12.00 | 12.00 | 11.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TCTGTTCTGCTTCCTCTGCAGt................................................. | 22 | t | 11.00 | 27.00 | 7.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - |
| ...................................................................................................................................................TTGTAGGTGAAGTAGACTTGGC................................................................................. | 22 | 1 | 11.00 | 11.00 | - | - | 1.00 | 4.00 | 1.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 |
| ...................................................................................................................................................................................TCTGTTCTGCTTCCTCTGCAGtt................................................ | 23 | tt | 10.00 | 27.00 | 7.00 | - | - | - | - | - | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TTTGTAGGTGAAGTAGACTTGG.................................................................................. | 22 | 1 | 9.00 | 9.00 | - | - | 3.00 | 1.00 | - | 1.00 | - | - | 1.00 | - | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - |
| ...................................................................................................................................................TTGTAGGTGAAGTAGACTTGG.................................................................................. | 21 | 1 | 7.00 | 7.00 | - | - | 1.00 | 1.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...................................................................................................................................................TTGTAGGTGAAGTAGACTTGGCa................................................................................ | 23 | a | 7.00 | 11.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TCTGTTCTGCTTCCTCTGCAtt................................................. | 22 | tt | 6.00 | 0.00 | - | 5.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TCTGTTCTGCTTCCTCTGCAGa................................................. | 22 | a | 3.00 | 27.00 | - | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TTGTAGGTGAAGTAGACTTGGCT................................................................................ | 23 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TCTGTTCTGCTTCCTCTGCcgtt................................................ | 23 | cgtt | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TTGTAGGTGAAGTAGACTTGGCTt............................................................................... | 24 | t | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................ATGAGGGGAAGCTGATGGCC.......... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TCTATGAGGGGAAGCTGATGGCCAGT....... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................................................................................................................................................................AGGGGAAGCTGATGGCCAG........ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TTGTAGGTGAAGTAGACTTGa.................................................................................. | 21 | a | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TTTGTAGGTGAAGTAGACTTGGCTGACAGt.......................................................................... | 30 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| AAAATCAATGCATTTAACAATGGTGCCATAGGGACCTTCTCTGTTATCACTGTCAGTGTTGTTAGGAAATCATTGGCGTCACAGGTATTCAGTGTTGGGACAAAATTAATTGTGGGCATAAATTAGTGTTGGAATTGGATGGAGTCTTTGTAGGTGAAGTAGACTTGGCTGACAGATGCTCTGTTCTGCTTCCTCTGCAGCTACACCTCTGAGCTTTTCTATGAGGGGAAGCTGATGGCCAGTGGGAAGC ....................................................................................................................................................((((((.(((((((((..(((........)))..)).))))))).))))))................................................... .............................................................................................................................................142.......................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesWT1() Testes Data. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................CAGATGCTCTGTTCTGata.............................................................. | 19 | ata | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |