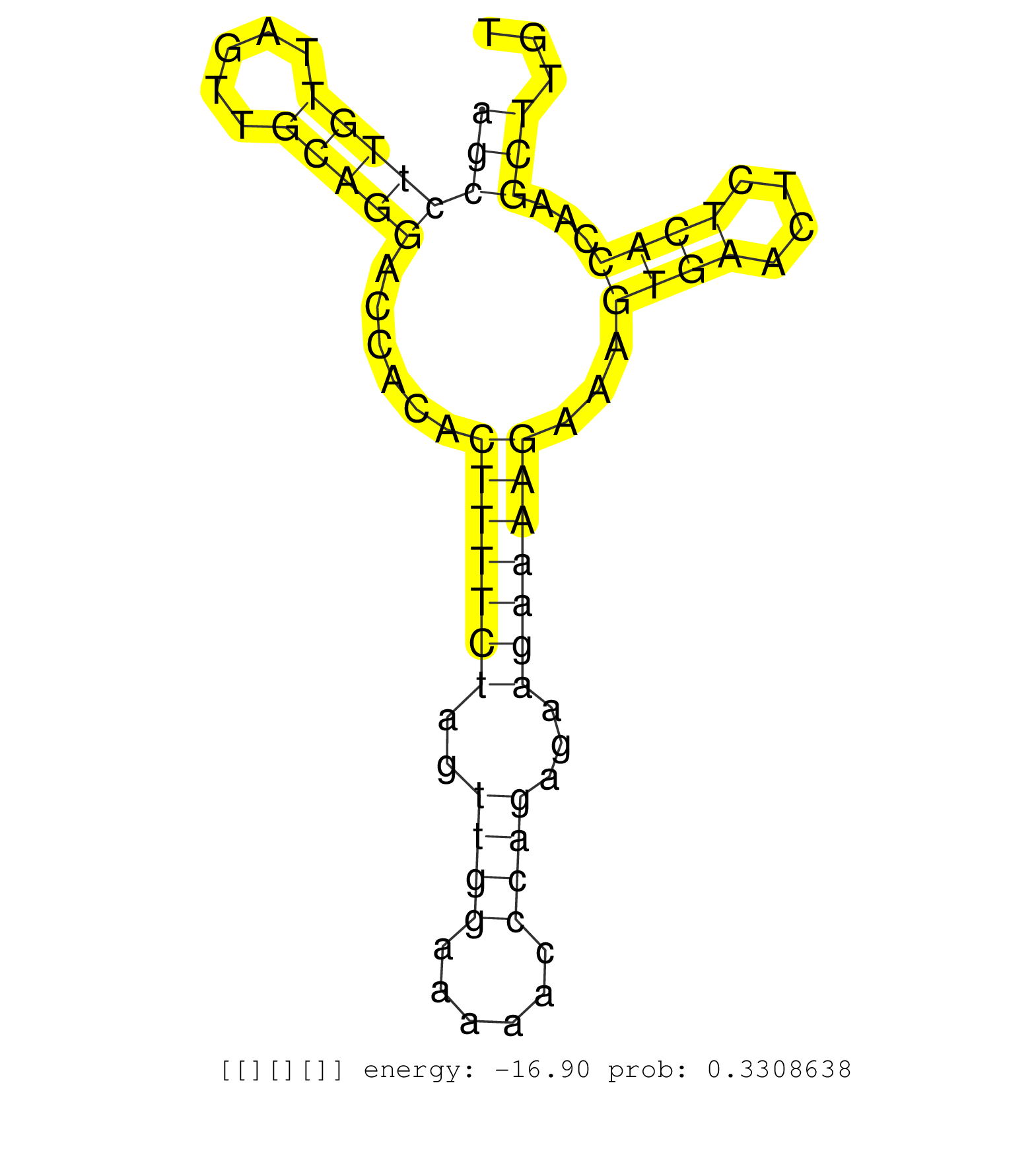

| Gene: Tcam1 | ID: uc007lyq.1_intron_5_0_chr11_106146956_f.5p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(21) TESTES |
| ACAGGGCCAGCGACTGGTCAGGACCACAGAGAGCCAGCTTCATGTCTTATGTGAGTAGAGGCCCGATCGCCCTGAACAAAGGCAAGATACTTAGTTTCCCTTACGGAGAGTTGGGTGGCAGCCTTGTTAGTTGCAGGACCACACTTTTCTAGTTGGAAAAACCCAGAGAAGAAAAGAAAGTGAACTCTCACCAAGCTTGTCTTGCCCTCAGGAAGGTGAGTGGGAGCAAGATGGGAGAGGCAGCCCAGAC .......................................................................................................................((((((((.....)))))......(((((((..((((......))))...)))))))...((((....))))...)))..................................................... .......................................................................................................................120.............................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR037903(GSM510439) testes_rep4. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................CAGAGAAGAAAAGAAAGTGAACTCTCACCAAGC...................................................... | 33 | 1 | 5.00 | 5.00 | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................TGTTAGTTGCAGGACCACACTTTTC..................................................................................................... | 25 | 1 | 4.00 | 4.00 | - | - | - | - | 1.00 | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................AAGAAAGTGAACTCTCACCAAGCTTGTC................................................. | 28 | 1 | 3.00 | 3.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .....................................................................................................................................................TAGTTGGAAAAACCCAGAGAAGAAAAGA......................................................................... | 28 | 1 | 3.00 | 3.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .....................................................................................................................................................TAGTTGGAAAAACCCAGAGAAGAAAAGAA........................................................................ | 29 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................AGTTGGAAAAACCCAGAGAAGAAAAGA......................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................GGCCCGATCGCCCTGAACAAAGGCAAG.................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................TCTTATGTGAGTAGAGGCCCGATCGCCC.................................................................................................................................................................................. | 28 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ........................................................................TGAACAAAGGCAAGATACTTAGTTTCCCTT.................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................TGAACAAAGGCAAGATACTTAGTTTCCCT..................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......CAGCGACTGGTCAGGACCACAGAGAGCC....................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .............CTGGTCAGGACCACAGAGAGCC....................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .....................................................AGTAGAGGCCCGATCGCCCTta............................................................................................................................................................................... | 22 | ta | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .............................................................................................................................................CACTTTTCTAGTTGGAAAAACCCAGAG.................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................AACCCAGAGAAGAAAAGAAAGTGAACTCTCtt........................................................... | 32 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .........................................................................................................................................................................AGAAAAGAAAGTGAACTCTCACCAAGCT..................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................AGTTGGAAAAACCCAGAGAAGAAAAaa......................................................................... | 27 | aa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .........................................................GAGGCCCGATCGCCCTGAACAAAGGCAA..................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TCAGGAAGGTGAGTGGGAGCAAGAT.................. | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................AGTAGAGGCCCGATCGCCCT................................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................AAGAAAGTGAACTCTCACCAAGCTTG................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................TTGTCTTGCCCTCAGGAAGGTGcgtg............................ | 26 | cgtg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................AACCCAGAGAAGAAAggat........................................................................ | 19 | ggat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................ACTTAGTTTCCCTTACGGAGAGTTGG........................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................TGTGAGTAGAGGCCCGATCGCCCTGA............................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........GCGACTGGTCAGGACC................................................................................................................................................................................................................................. | 16 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - |
| ACAGGGCCAGCGACTGGTCAGGACCACAGAGAGCCAGCTTCATGTCTTATGTGAGTAGAGGCCCGATCGCCCTGAACAAAGGCAAGATACTTAGTTTCCCTTACGGAGAGTTGGGTGGCAGCCTTGTTAGTTGCAGGACCACACTTTTCTAGTTGGAAAAACCCAGAGAAGAAAAGAAAGTGAACTCTCACCAAGCTTGTCTTGCCCTCAGGAAGGTGAGTGGGAGCAAGATGGGAGAGGCAGCCCAGAC .......................................................................................................................((((((((.....)))))......(((((((..((((......))))...)))))))...((((....))))...)))..................................................... .......................................................................................................................120.............................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR037903(GSM510439) testes_rep4. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................AACCCAGAGAAGAAAatc............................................................................ | 18 | atc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |