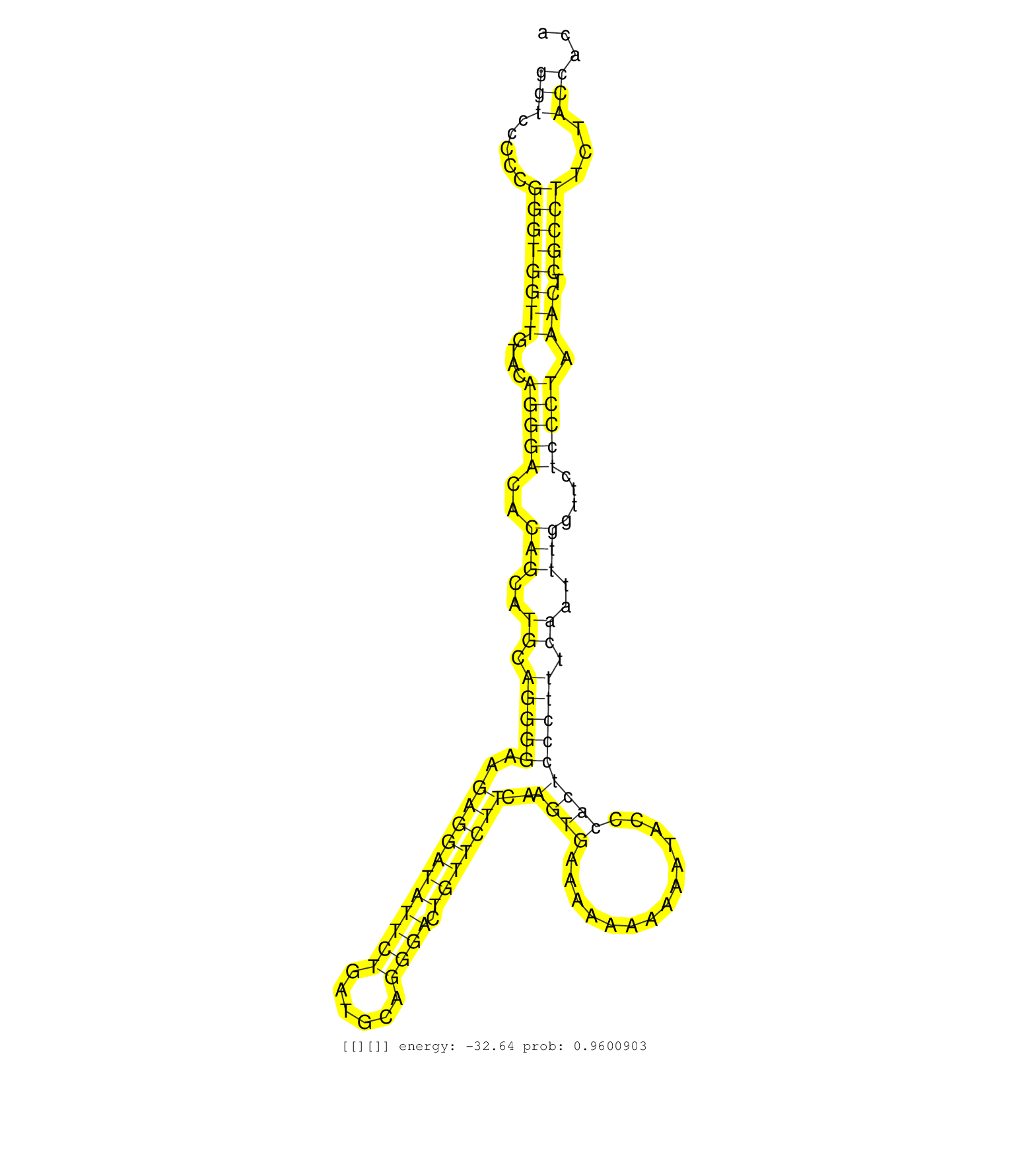

| Gene: Ace | ID: uc007lxu.1_intron_20_0_chr11_105847032_f | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(2) PIWI.ip |
(1) PIWI.mut |
(9) TESTES |
| GATGGAAGCATCACCAAGGAGAACTATAACCAGGAGTGGTGGAGCCTCAGGTTCTAGAACACTCTGGTGGGGGTGTGGATTGGAAGGGATCTATGAATTGTCTGCATGCGCCTTGAGGGTCTACAGATGAGCCAGTAAAGTGTATGTGTGCGTACACACCAGTGATGGGGGGTCCCCCGGGTGGTTGTACAGGGACACAGCATGCAGGGGAAGAGGATATTCTGATGCAGGGACTGTTCTTCAAGTGAAAAAAAAAATACCCACTCCCTTTCAATTTGGTTCTCCCTAAACTCGCCTTCTACCACATCTTCCTCCCCAACAGGCTGAAGTATCAGGGTCTGTGCCCCCCAGTGCCAAGATCCCAAGGTGACT ..........................................................................................................................................................................(((.....((((((((....(((((..(((..((.(((((..(((((((((((......)))).)))))))..((((..............))))))))).))..)))....))))).))).)))))...)))..................................................................... ..........................................................................................................................................................................171....................................................................................................................................306................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................CCCGGGTGGTTGTACAGGGACACAGCATGCAGGGGAAGAGGATATTCTGATG................................................................................................................................................. | 52 | 1 | 59.00 | 59.00 | 59.00 | - | - | - | - | - | - | - | - |
| ..............................CAGGAGTGGTGGAGCCTCAGGctg.............................................................................................................................................................................................................................................................................................................................. | 24 | ctg | 6.00 | 0.00 | - | 6.00 | - | - | - | - | - | - | - |
| .............CCAAGGAGAACTATAACCAGGAGTGGT............................................................................................................................................................................................................................................................................................................................................ | 27 | 1 | 4.00 | 4.00 | - | 4.00 | - | - | - | - | - | - | - |
| ...........CACCAAGGAGAACTATAACCAGGAG................................................................................................................................................................................................................................................................................................................................................ | 25 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - |
| ......AGCATCACCAAGGAGAACTATAACCAGGAGTGGT............................................................................................................................................................................................................................................................................................................................................ | 34 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - |
| ............................................................................................................................................................................................................................................................................................CCTAAACTCGCCTTaaga...................................................................... | 18 | aaga | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - |
| ..........TCACCAAGGAGAACTATAACCAGGAG................................................................................................................................................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - |
| ..............................................................................ATTGGAAGGGATCTATGAATTGTCTGCATGCGC..................................................................................................................................................................................................................................................................... | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - |
| ..TGGAAGCATCACCAAGGAGAACTATAAC...................................................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - |
| ................AGGAGAACTATAACCAGGAGTGGTGGA......................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................................................................................................................................AGTATCAGGGTCTGT.............................. | 15 | 4 | 0.25 | 0.25 | - | - | 0.25 | - | - | - | - | - | - |
| GATGGAAGCATCACCAAGGAGAACTATAACCAGGAGTGGTGGAGCCTCAGGTTCTAGAACACTCTGGTGGGGGTGTGGATTGGAAGGGATCTATGAATTGTCTGCATGCGCCTTGAGGGTCTACAGATGAGCCAGTAAAGTGTATGTGTGCGTACACACCAGTGATGGGGGGTCCCCCGGGTGGTTGTACAGGGACACAGCATGCAGGGGAAGAGGATATTCTGATGCAGGGACTGTTCTTCAAGTGAAAAAAAAAATACCCACTCCCTTTCAATTTGGTTCTCCCTAAACTCGCCTTCTACCACATCTTCCTCCCCAACAGGCTGAAGTATCAGGGTCTGTGCCCCCCAGTGCCAAGATCCCAAGGTGACT ..........................................................................................................................................................................(((.....((((((((....(((((..(((..((.(((((..(((((((((((......)))).)))))))..((((..............))))))))).))..)))....))))).))).)))))...)))..................................................................... ..........................................................................................................................................................................171....................................................................................................................................306................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................GCCTTGAGGGTCTACAGATGAGCCAGT............................................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - |
| .........................................................................................................ATGCGCCTTGAGGGTCTACAGATGAGCCA.............................................................................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 |