
| Gene: Tlk2 | ID: uc007lxd.1_intron_4_0_chr11_105071152_f.5p | SPECIES: mm9 |
 |
 |
|
(2) PIWI.ip |
(8) TESTES |
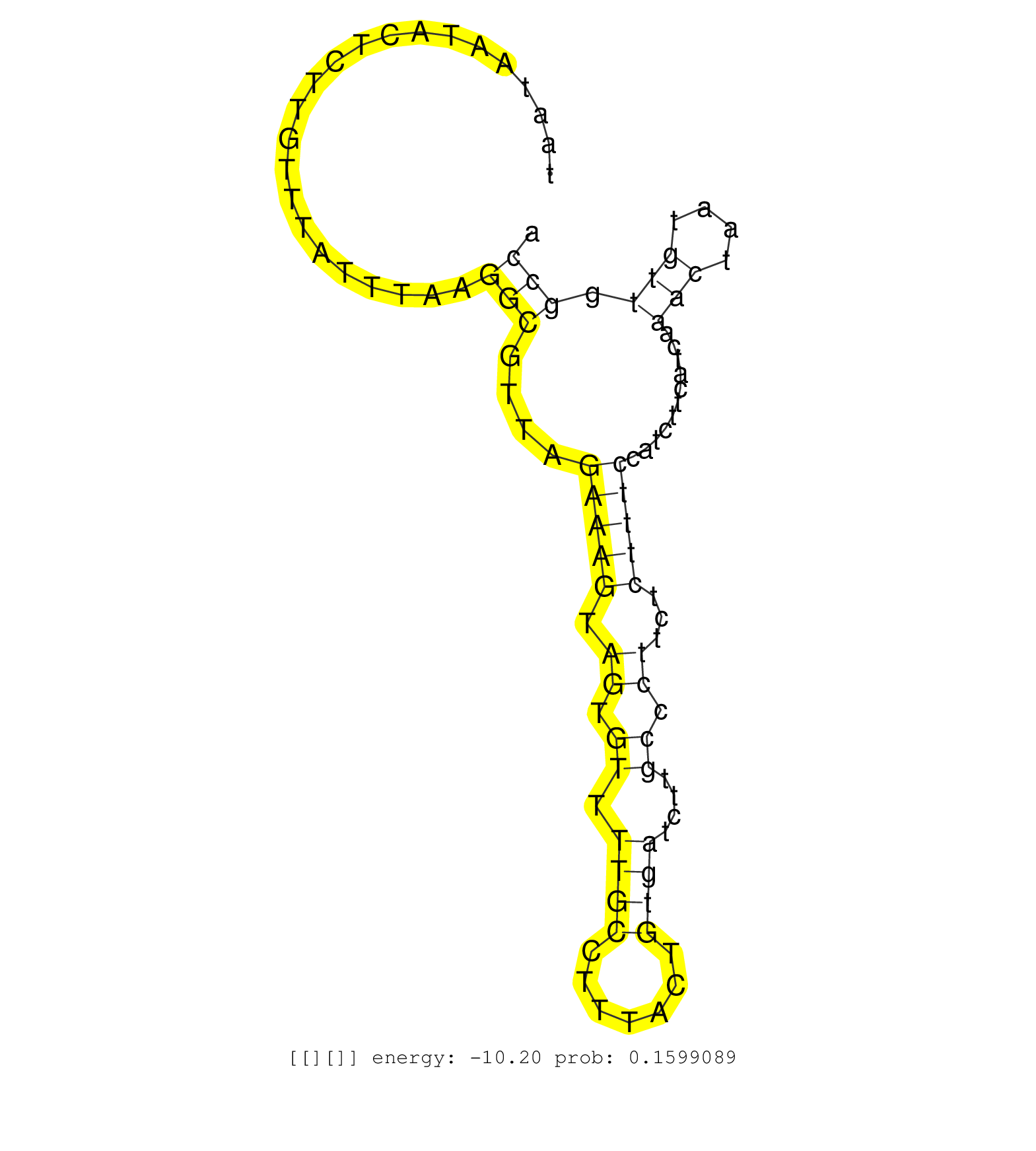
| TTTCAGGGAAAGGCACTCCTAGGGGACATAAAATTAGTGATTACTTTGAGGTAAGTTAAATTGGGGGTTAAAAAACAACCTCCAGTATCTTTAAACAACTTAATAATACTCTTGTTTATTTAAGGCGTTAGAAAGTAGTGTTTTGCCTTTACTGTGATCTTGCCCTTCTCTTTCCATCTTCATCAAACTAATGTTGGCCATCTTGTAATTAGAAGGACTGCTGCAGGTTTGCTAGGTTTAAGTTTTGTAT ...........................................................................................................................(((....(((((.((.((.((((.......))))....)).))...)))))...........(((....))).)))................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................AATACTCTTGTTTATTTAAGGCGTTAGAAAGTAGTGTTTTGCCTTTACTGTG.............................................................................................. | 52 | 1 | 24.00 | 24.00 | 24.00 | - | - | - | - | - | - | - |
| ............................TAAAATTAGTGATTACTTTGAGcgtc.................................................................................................................................................................................................... | 26 | cgtc | 6.00 | 0.00 | - | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - |
| ....................AGGGGACATAAAATTAGTGATTACTTTGAG........................................................................................................................................................................................................ | 30 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - |
| ...................TAGGGGACATAAAATTAGTGATTACTTTGAGt....................................................................................................................................................................................................... | 32 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - |
| .......................GGACATAAAATTAGTGATTACTTTGAG........................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - |
| ...................TAGGGGACATAAAATTAGTGATTACTTTGt......................................................................................................................................................................................................... | 30 | t | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - |
| ..............................AAATTAGTGATTACTTTGAGcgtc.................................................................................................................................................................................................... | 24 | cgtc | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - |
| ...................TAGGGGACATAAAATTAGTGATTACTTTG.......................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 |
| ....................AGGGGACATAAAATTAGTGATTACTTT........................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - |
| ...................TAGGGGACATAAAATTAGTGATTACTTTGA......................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - |
| ..................CTAGGGGACATAAAATTAGTGATTACTTTG.......................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - |
| ...................TAGGGGACATAAAATTAGTGATTACTTT........................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - |
| TTTCAGGGAAAGGCACTCCTAGGGGACATAAAATTAGTGATTACTTTGAGGTAAGTTAAATTGGGGGTTAAAAAACAACCTCCAGTATCTTTAAACAACTTAATAATACTCTTGTTTATTTAAGGCGTTAGAAAGTAGTGTTTTGCCTTTACTGTGATCTTGCCCTTCTCTTTCCATCTTCATCAAACTAATGTTGGCCATCTTGTAATTAGAAGGACTGCTGCAGGTTTGCTAGGTTTAAGTTTTGTAT ...........................................................................................................................(((....(((((.((.((.((((.......))))....)).))...)))))...........(((....))).)))................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|