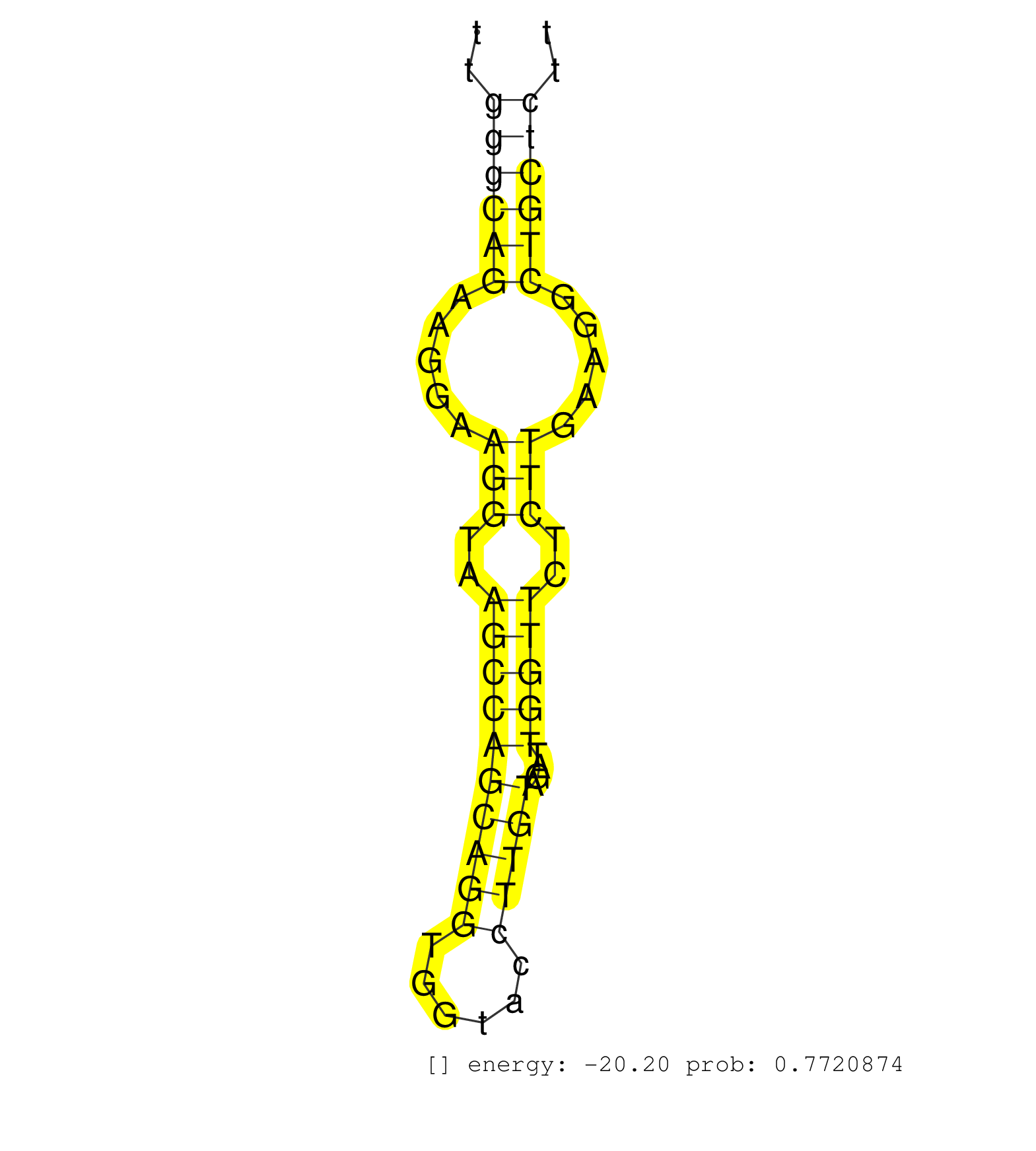
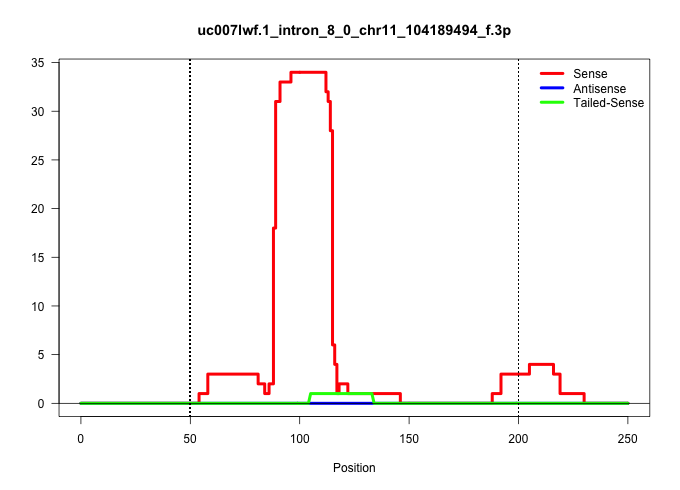
| Gene: Mapt | ID: uc007lwf.1_intron_8_0_chr11_104189494_f.3p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(4) PIWI.ip |
(11) TESTES |
| GTAACGTGGGTGTGTGTGCCCCTTCTGCAGGGCAGCCTGTGGGAGAAGGGGTATTGGGCAGAAGGAAGGTAAGCCAGCAGGTGGTACCTTGTAGATTGGTTCTCTTGAAGGCTGCTCTTGACATCCCAGGGCACTGGCTTCTTCCTCCCTCCCCGCAAGGTGGGAGGTCCTGAGCGAGGTGTTTCCCTTCGCTCCCACAGGAAAAGCTGCTTTACTGAGTTCTCAAGTTTGGAACTACAGCCATGATTTG .......................................................((((((.....(((..((((((((((......)))))....)))))..))).....))))))..................................................................................................................................... .....................................................54...............................................................119................................................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................TTGTAGATTGGTTCTCTTGAAGGCTGC....................................................................................................................................... | 27 | 1 | 12.00 | 12.00 | 5.00 | 1.00 | 3.00 | 2.00 | - | - | 1.00 | - | - | - | - |
| .........................................................................................TGTAGATTGGTTCTCTTGAAGGCTGC....................................................................................................................................... | 26 | 1 | 11.00 | 11.00 | 3.00 | 2.00 | 1.00 | - | 1.00 | 2.00 | 1.00 | - | - | - | 1.00 |
| ........................................................................................TTGTAGATTGGTTCTCTTGAAGGCTG........................................................................................................................................ | 26 | 1 | 3.00 | 3.00 | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................................TAGATTGGTTCTCTTGAAGGCTGCTC..................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................TGTAGATTGGTTCTCTTGAAGGCTGCT...................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ..........................................................CAGAAGGAAGGTAAGCCAGCAGGTGG...................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................TCCCACAGGAAAAGCTGCTTTACTGAG............................... | 27 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................GCTGCTTTACTGAGTTCTCAAGTTT.................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .........................................................................................................TGAAGGCTGCTCTTGACATCCCAGGGCAt.................................................................................................................... | 29 | t | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................................................................................TTGTAGATTGGTTCTCTTGAAGGC.......................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ......................................................................................CCTTGTAGATTGGTTCTCTTGAAGGC.......................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................TGTAGATTGGTTCTCTTGAAGGCTGCTC..................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................TACCTTGTAGATTGGTTCTCTTGAAGGCT......................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......................................................................................................................TGACATCCCAGGGCACTGGCTTCTTCCT........................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................TGGGCAGAAGGAAGGTAAGCCAGCAGG......................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................................................................................................................................................TCGCTCCCACAGGAAAAGCTGCTTTACT.................................. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................TGGTTCTCTTGAAGGCTGCTCTTGAC................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .........................................................................................TGTAGATTGGTTCTCTTGAAGGCTG........................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| GTAACGTGGGTGTGTGTGCCCCTTCTGCAGGGCAGCCTGTGGGAGAAGGGGTATTGGGCAGAAGGAAGGTAAGCCAGCAGGTGGTACCTTGTAGATTGGTTCTCTTGAAGGCTGCTCTTGACATCCCAGGGCACTGGCTTCTTCCTCCCTCCCCGCAAGGTGGGAGGTCCTGAGCGAGGTGTTTCCCTTCGCTCCCACAGGAAAAGCTGCTTTACTGAGTTCTCAAGTTTGGAACTACAGCCATGATTTG .......................................................((((((.....(((..((((((((((......)))))....)))))..))).....))))))..................................................................................................................................... .....................................................54...............................................................119................................................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) |
|---|