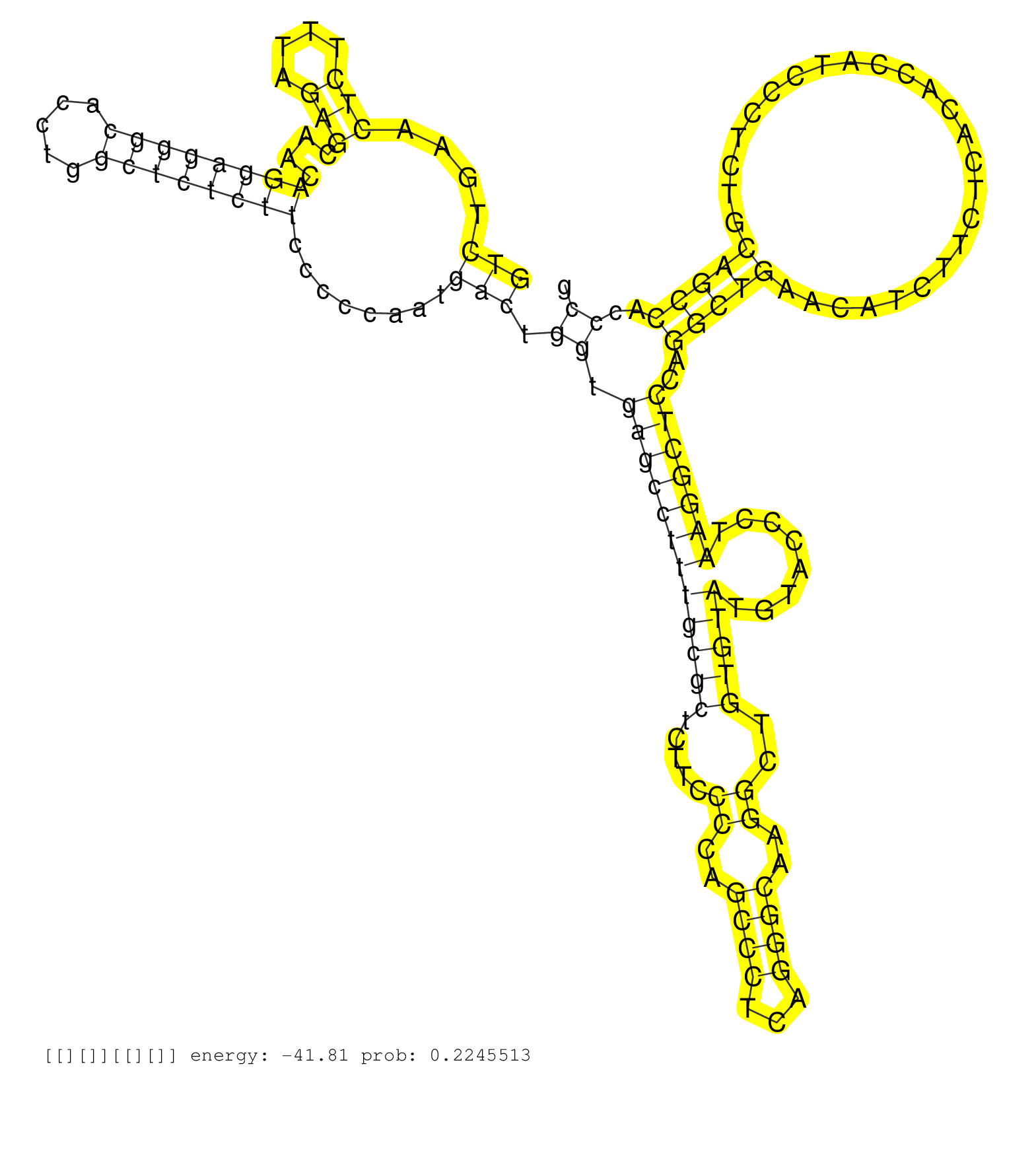
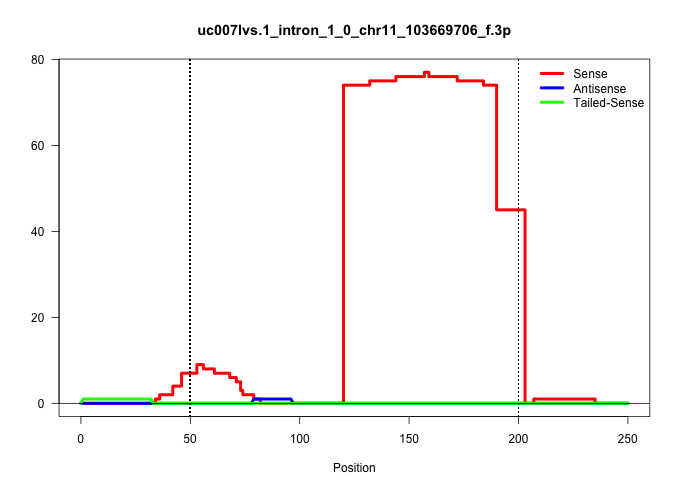
| Gene: Wnt3 | ID: uc007lvs.1_intron_1_0_chr11_103669706_f.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(2) PIWI.ip |
(1) PIWI.mut |
(13) TESTES |
| CTGCAGCTCTGAACTCAGCTTGCCCATCCACAAAGAGAATACTAACTAGGGTCTGAACTCTTTAGAGCAACAGGAGGGCACCTGGCTCTCTTCCCCCAATGACTGGTGAGCCTTTGCGCTCTTCCCCAGCCCTCAGGGCAAGGCTGTGTATGTACCCTAAGGCTCCAGGCTGAACATCTTCTCACACCATCCCTCTGCAGCCACCCGTGAATCGGCCTTCGTGCATGCCATCGCCTCGGCTGGTGTCGCC ..................................................(((....(((....)))....((((((((.....))))))))........))).((.((((((((((((.....((..((((...))))..))..)))))........)))))))..(((((.........................)))))..))............................................ ..................................................51..........................................................................................................................................................207......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT4() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................CTTCCCCAGCCCTCAGGGCAAGGCTGTGTATGTACCCTAAGGCTCCAGGCTG.............................................................................. | 52 | 1 | 75.00 | 75.00 | 75.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................TGAACTCTTTAGAGCAACAGGAGGGC........................................................................................................................................................................... | 26 | 1 | 5.00 | 5.00 | - | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................TAGGGTCTGAACTCTTTAGAGCAACAGG................................................................................................................................................................................ | 28 | 1 | 4.00 | 4.00 | - | - | - | 1.00 | 3.00 | - | - | - | - | - | - | - | - |
| ..............................................TAGGGTCTGAACTCTTTAGAGCAACAG................................................................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................GAGAATACTAACTAGGGTCTGA.................................................................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..........................................TAACTAGGGTCTGAACTCTTTAGAGC...................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TGTGTATGTACCCTAAGGCTCCAGGCTG.............................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .............................................................................................................................................................TAAGGCTCCAGGCTGAACATCTTCTCA.................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...............................................................................................................................................................................................................TGAATCGGCCTTCGTGCATGCCATCGCC............... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .....................................................TGAACTCTTTAGAGCAACAGGAGGGCACC........................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ....................................GAATACTAACTAGGGTCTGAACTCT............................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .TGCAGCTCTGAACTCAGCTTGCCCATCCACAt......................................................................................................................................................................................................................... | 32 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TCAGGGCAAGGCTGTGTATGTACCCTA........................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........................................TAACTAGGGTCTGAACTCTTTAGAGCAAC................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| CTGCAGCTCTGAACTCAGCTTGCCCATCCACAAAGAGAATACTAACTAGGGTCTGAACTCTTTAGAGCAACAGGAGGGCACCTGGCTCTCTTCCCCCAATGACTGGTGAGCCTTTGCGCTCTTCCCCAGCCCTCAGGGCAAGGCTGTGTATGTACCCTAAGGCTCCAGGCTGAACATCTTCTCACACCATCCCTCTGCAGCCACCCGTGAATCGGCCTTCGTGCATGCCATCGCCTCGGCTGGTGTCGCC ..................................................(((....(((....)))....((((((((.....))))))))........))).((.((((((((((((.....((..((((...))))..))..)))))........)))))))..(((((.........................)))))..))............................................ ..................................................51..........................................................................................................................................................207......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT4() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................GGGTCTGAACTCTTacca............................................................................................................................................................................................ | 18 | acca | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................ACCTGGCTCTCTTCCCCC......................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................TGAACATCTTCTCAatg.................................................................. | 17 | atg | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |