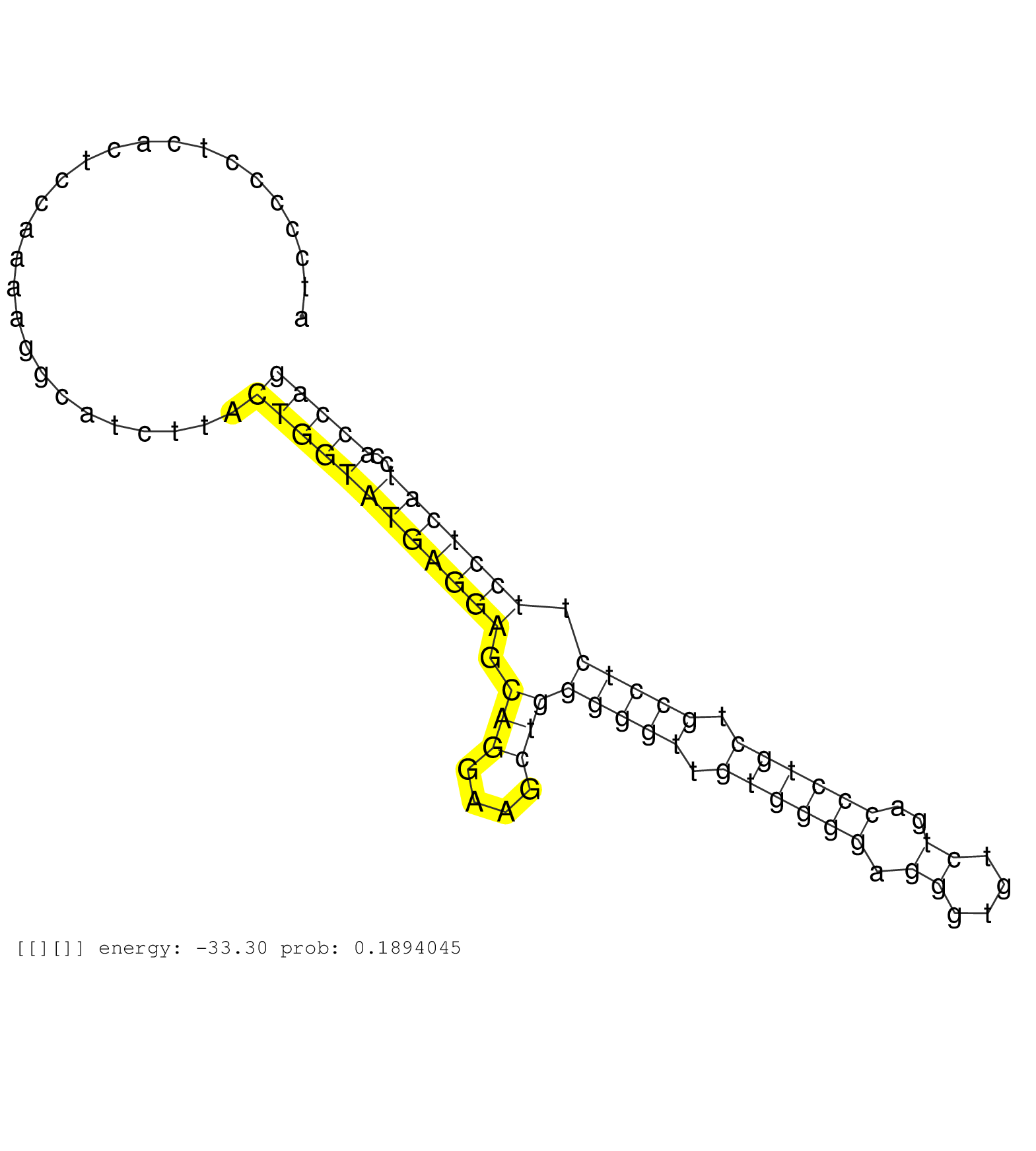

| Gene: Fr1 | ID: uc007ltw.1_intron_0_0_chr11_103046653_f | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(5) TESTES |
| GTTTTGAGTTCCAGTCCTCTGCCCTTACTTCCCAAATACTGGAATTACAGGCATGTGCCATTGTGCCTGGCTCACAGCAGCCCTTTTAAAAAACCAAGTAATTTTAAGGGCCTTTGCTTCCTTGGCCCACCCTCACTCCCTCCCCCAGCATGCCTGCCCCTGGGGAAGGCAGCCTTCAGCTGGCCAGCTGAGTCAGGTAGATAGGAGACAGCAGCTTAGAGTTCCTAGATCGATCCCCCTCACTCCAAAAGGCATCTTACTGGTATGAGGAGCAGGAAGCTGGGGGTTGTGGGGAGGGTGTCTGACCCTGCTGCCTCTTCCTCATCCACCAGGCTGACCCCTGCCCACAGCAGGAAGGCGCTGAGGAATTCCCGAATCGTCA ...................................................................................................................................................................................................................................................................((((((((((((.(((....)))(((((.((((((.((....))..)))))).))))).)))))))..))))).................................................. ........................................................................................................................................................................................................................................233................................................................................................332................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................................................................................................ACTGGTATGAGGAGCAGGAAG....................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - |
| GTTTTGAGTTCCAGTCCTCTGCCCTTACTTCCCAAATACTGGAATTACAGGCATGTGCCATTGTGCCTGGCTCACAGCAGCCCTTTTAAAAAACCAAGTAATTTTAAGGGCCTTTGCTTCCTTGGCCCACCCTCACTCCCTCCCCCAGCATGCCTGCCCCTGGGGAAGGCAGCCTTCAGCTGGCCAGCTGAGTCAGGTAGATAGGAGACAGCAGCTTAGAGTTCCTAGATCGATCCCCCTCACTCCAAAAGGCATCTTACTGGTATGAGGAGCAGGAAGCTGGGGGTTGTGGGGAGGGTGTCTGACCCTGCTGCCTCTTCCTCATCCACCAGGCTGACCCCTGCCCACAGCAGGAAGGCGCTGAGGAATTCCCGAATCGTCA ...................................................................................................................................................................................................................................................................((((((((((((.(((....)))(((((.((((((.((....))..)))))).))))).)))))))..))))).................................................. ........................................................................................................................................................................................................................................233................................................................................................332................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................................................................................................................................................................TTCCTCATCCACCAGGCcgta................................................ | 21 | cgta | 1.00 | 0.00 | 1.00 | - | - | - | - |
| ..........................................................................................................................................................................................................................................................TCTTACTGGTATGAGctga................................................................................................................. | 19 | ctga | 1.00 | 0.00 | - | - | - | 1.00 | - |
| .....................................................................ACAGCAGCCCTTTTAaggc...................................................................................................................................................................................................................................................................................................... | 19 | aggc | 1.00 | 0.00 | - | - | 1.00 | - | - |
| ................................................................................................................................................................................................................................................................................................................CTGCTGCCTCTTCCTCAaag.......................................................... | 20 | aag | 1.00 | 0.00 | - | - | - | - | 1.00 |