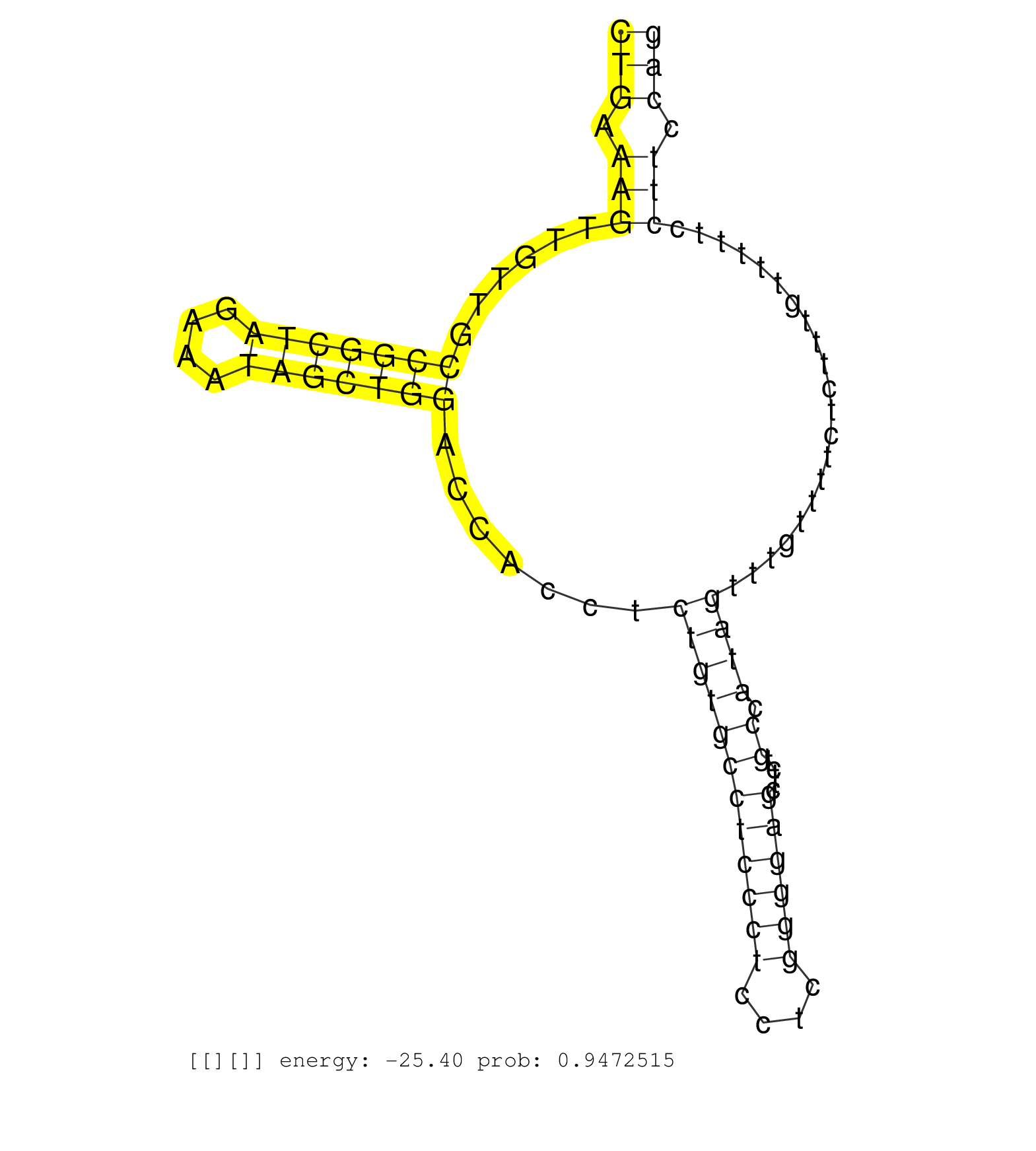

| Gene: mKIAA0031 | ID: uc007lss.1_intron_0_0_chr11_102730485_r.3p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(3) PIWI.ip |
(1) PIWI.mut |
(16) TESTES |
| ATACTTTCATCCAGAGGGCACTGGTCTCATGATCTTGGTTTCCAGCCTGTGTTGTCTTGTGAGCACCCTGACCTAGGGCTCTGAATGGTCTGCAGTTGATCTGAAAGTTGTTGCCGGCTAGAAATAGCTGGACCACCTCTGTGCCTCCCTCCTCGGGGAGCTTCTGCCATAGTTTGTTTTCTCTTTGTTTTTCCTTCCAGTTTCTTAGCAGATCTGATGGACAACTCTGAGCTCATCAGAAACGTAACCT ....................................................................................................(((.(((......(((((((....))))))).......((((((((((((....)))))).....)).)))).....................))).))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................GAGCACCCTGACCTAGGGCTCTGAATGGTCTGCAGTTGATCTGAAAGTTGTT.......................................................................................................................................... | 52 | 1 | 31.00 | 31.00 | 31.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TGGACAACTCTGAGCTCATCAGAAACGT..... | 28 | 1 | 4.00 | 4.00 | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .......................................................................CCTAGGGCTCTGAATGG.................................................................................................................................................................. | 17 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TGGACAACTCTGAGCTCATCAGAAACGTA.... | 29 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................GTTTCTTAGCAGATCTGATGGACAACTCTGAG................... | 32 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TTCTTAGCAGATCTGATGGACAACTCT...................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .........................................................................................................................................................................................................................TGGACAACTCTGAGCTCATCAGAA......... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TGGACAACTCTGAGCTCATCAGAAACG...... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TAGCAGATCTGATGGACAACTCTGAG................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................CAACTCTGAGCTCATCAGAAACGTAACCT | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................TTTCTTAGCAGATCTGATGGACAACTC....................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................ATCTGATGGACAACTCTGAGCTCATCAGAAA........ | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................................................................................................................................................................................................TTCTTAGCAGATCTGATGGAC............................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................GACAACTCTGAGCTCATCAGAAAC....... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..........................................................................................................................................................................................................TCTTAGCAGATCTGATGGACAACTCT...................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................ACAACTCTGAGCTCATCAGAAACGTAAac. | 29 | ac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..............................................................................................GTTGATCTGAAAGTTGTTGCCGGCTA.................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................ATGGACAACTCTGAGt.................. | 16 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TAGCAGATCTGATGGACAACTCTGAGC.................. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................GAGCTCATCAGAAACGTAACCT | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................................................................................................................................................................................................TTAGCAGATCTGATGGACAA.......................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TAGCAGATCTGATGGACAACTCTaagc.................. | 27 | aagc | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................TGAGCTCATCAGAAACGTAA... | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ATACTTTCATCCAGAGGGCACTGGTCTCATGATCTTGGTTTCCAGCCTGTGTTGTCTTGTGAGCACCCTGACCTAGGGCTCTGAATGGTCTGCAGTTGATCTGAAAGTTGTTGCCGGCTAGAAATAGCTGGACCACCTCTGTGCCTCCCTCCTCGGGGAGCTTCTGCCATAGTTTGTTTTCTCTTTGTTTTTCCTTCCAGTTTCTTAGCAGATCTGATGGACAACTCTGAGCTCATCAGAAACGTAACCT ....................................................................................................(((.(((......(((((((....))))))).......((((((((((((....)))))).....)).)))).....................))).))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|