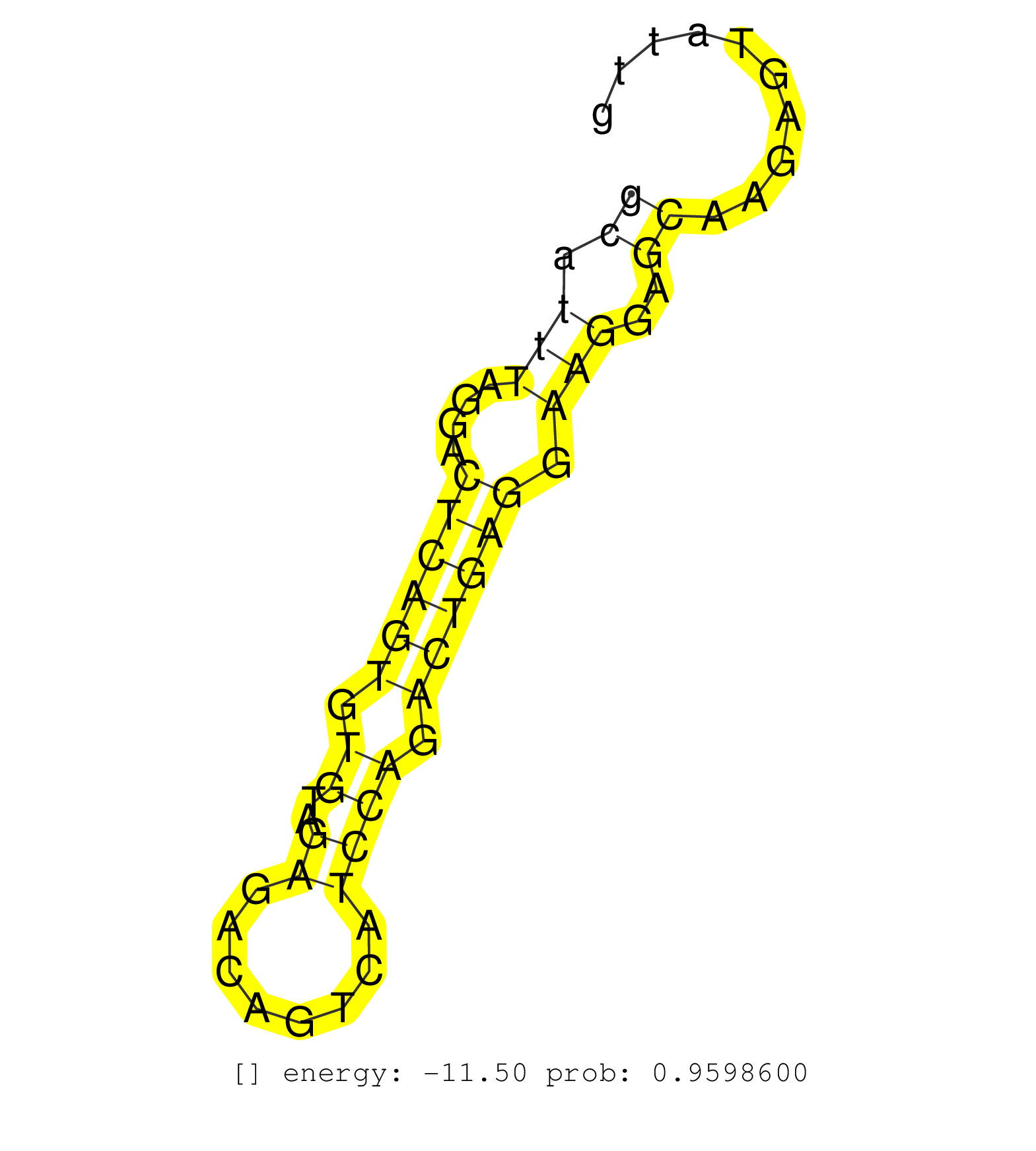
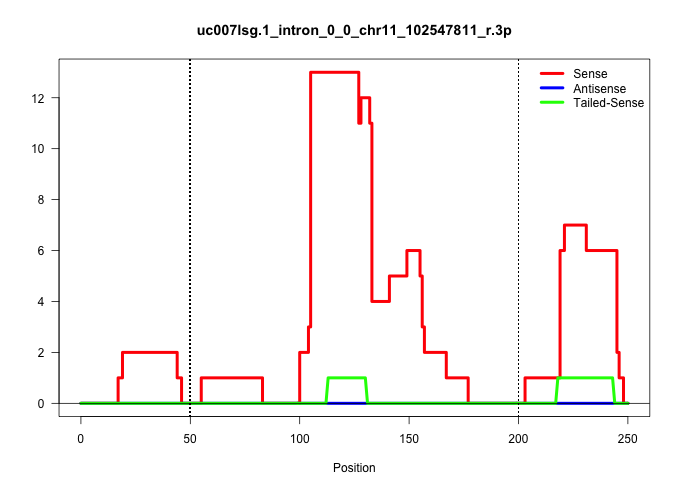
| Gene: Ccdc43 | ID: uc007lsg.1_intron_0_0_chr11_102547811_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(18) TESTES |
| TCTGAACCCAGGTCCTTTCTGTAAGAGCAAGTGCCTATGACCACTGAGCCATCTCTAGTCCAAACCAAAGATGCTTTCCTGTACTAGGAATAGCTAGAGTGCATTTAGGACTCAGTGTGTAGAGACAGTCATCCAGACTGAGGAAGGAGCAAGAGTATTGCCTACAGGACTTGGGCCTGAATCCCTTAACTCACTGGTAGCCTTGTTCCGAAACACCAATGTGGAAGATGTTCTCAATGCTCGGAAATTG ....................................................................................................((.(((....((((((.((..((........)))).)))))).)))..)).................................................................................................... ....................................................................................................101........................................................160........................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................TCCAGACTGAGGAAGGAGCAAGAGT.............................................................................................. | 25 | 1 | 10.00 | 10.00 | 6.00 | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .........................................................................................................TAGGACTCAGTGTGTAGAGACAGTCATC..................................................................................................................... | 28 | 1 | 7.00 | 7.00 | - | 4.00 | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................TGTGGAAGATGTTCTCAATGCTCGGA..... | 26 | 1 | 5.00 | 5.00 | - | - | 1.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TAGGACTCAGTGTGTAGAGACAGTCA....................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................GCATTTAGGACTCAGTGTGTAGAGACA........................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................TGTGGAAGATGTTCTCAATGCTCGGAA.... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................TGTAAGAGCAAGTGCCTATGACCACTG............................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TAGGACTCAGTGTGTAGAGACAGTCAT...................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................TCCAGACTGAGGAAGGAGCAAGAGTA............................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................TGTGGAAGATGTTCTCAATGCTCG....... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................................................................................................................................................................TGTTCCGAAACACCAATGTGGAAGATGT................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .....................................................................................................................................................CAAGAGTATTGCCTACAGGACTTGGGCC......................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................TCTGTAAGAGCAAGTGCCTATGACCAC.............................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................ATCCAGACTGAGGAAaaaa..................................................................................................... | 19 | aaaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .............................................................................................................................................................................................................................TGGAAGATGTTCTCAATGCTCGGAAAT.. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................GGAAGGAGCAAGAGTATTGCCTACAG................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................ATGTGGAAGATGTTCTCAATGCTCGt...... | 26 | t | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................TCATCCAGACTGAGGAAGGAGCAAGAG............................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................TTAGGACTCAGTGTGTAGAGACAGTCA....................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................AGTGTGTAGAGACAcacc....................................................................................................................... | 18 | cacc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .......................................................TAGTCCAAACCAAAGATGCTTTCCTGTA....................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| TCTGAACCCAGGTCCTTTCTGTAAGAGCAAGTGCCTATGACCACTGAGCCATCTCTAGTCCAAACCAAAGATGCTTTCCTGTACTAGGAATAGCTAGAGTGCATTTAGGACTCAGTGTGTAGAGACAGTCATCCAGACTGAGGAAGGAGCAAGAGTATTGCCTACAGGACTTGGGCCTGAATCCCTTAACTCACTGGTAGCCTTGTTCCGAAACACCAATGTGGAAGATGTTCTCAATGCTCGGAAATTG ....................................................................................................((.(((....((((((.((..((........)))).)))))).)))..)).................................................................................................... ....................................................................................................101........................................................160........................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|