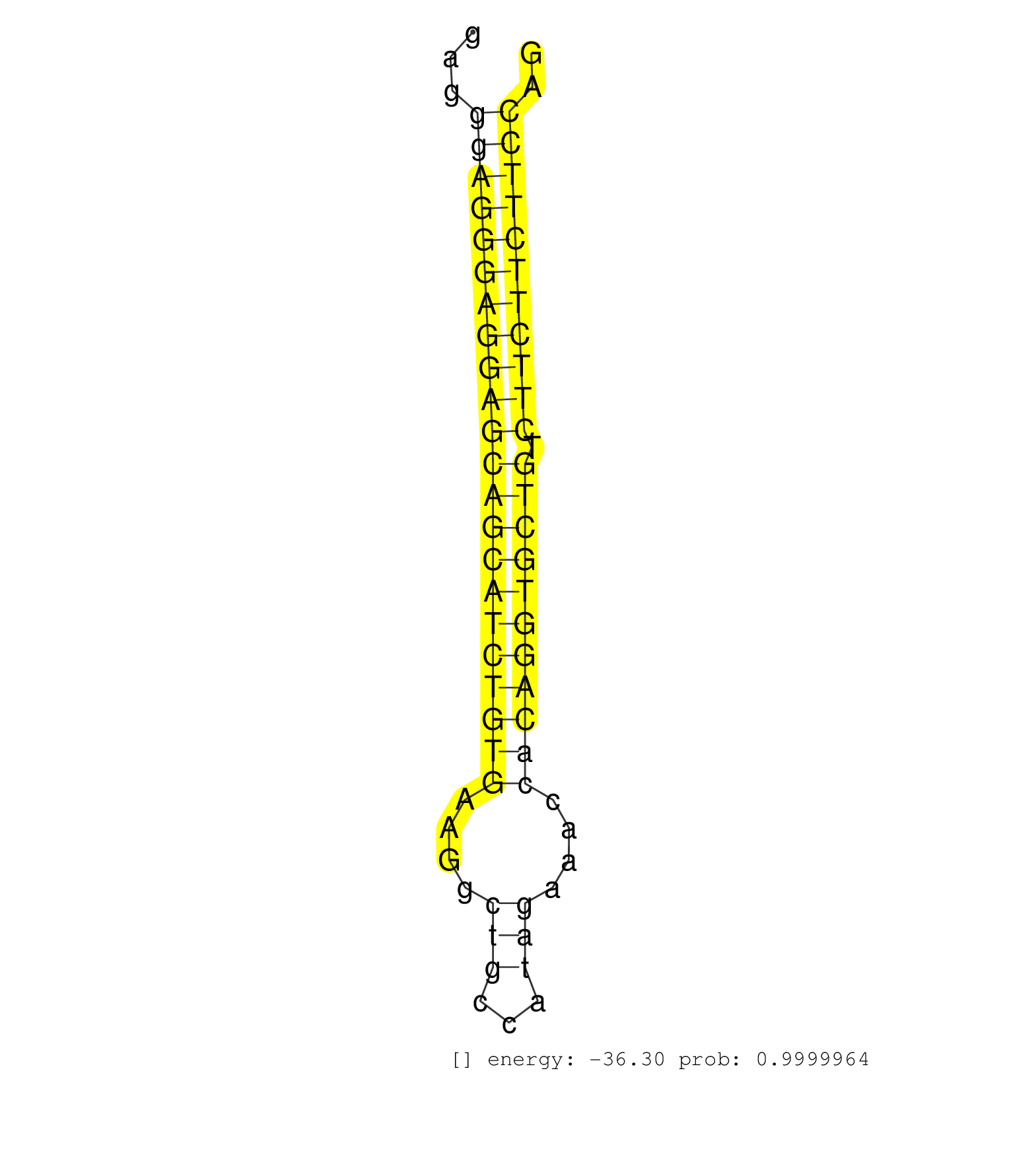
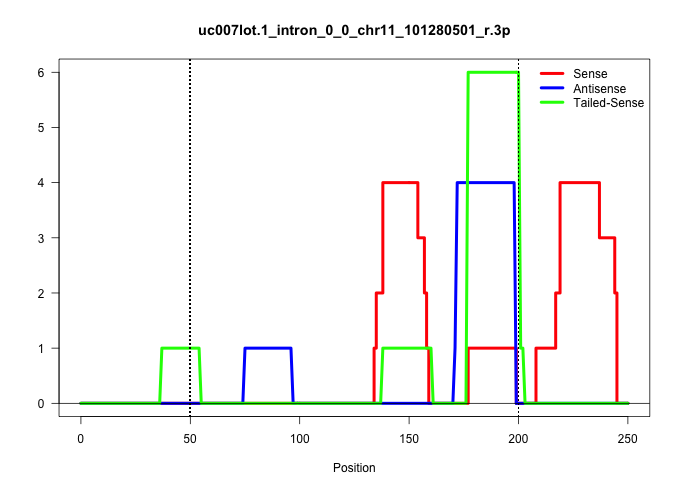
| Gene: 1700113I22Rik | ID: uc007lot.1_intron_0_0_chr11_101280501_r.3p | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(1) OVARY |
(5) PIWI.ip |
(1) PIWI.mut |
(21) TESTES |
| GTGACACCGTTTGAAATCCTCAAAACAAGGGTGCACAATAGAGTTCTGTTCACTGAGTCCTGGTGCTTTACCTCATTCCCCAAAGTGAGCCTCCCAAGAGATGTGACTCGGGATTCCAGGAGAGCAGCTGGGGGAGGGAGGGAGGAGCAGCATCTGTGAAGGCTGCCATAGAAACCACAGGTGCTGTCTTCTTCTTCCAGGATGATTCTGACAGCTAACTAGCTTTCTGTGACGGTGGAGCCGGGGAGGA ........................................................................................................................................((((((((((((((((((((((....(((...)))....))))))))))).))))))))))).................................................... .....................................................................................................................................134...............................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT4() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................TGACTCGGGATTCCAGGAGAGCAGCTGGGGGAGGGAGGGAGGAGCAGCATCT............................................................................................... | 52 | 1 | 16.00 | 16.00 | 16.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................CAGGTGCTGTCTTCTTCTTCCAGat................................................ | 25 | at | 8.00 | 0.00 | - | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................CAGGTGCTGTCTTCTTCTTCCAGt................................................. | 24 | t | 7.00 | 0.00 | - | - | - | - | - | 1.00 | 2.00 | 1.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................TAGCTTTCTGTGACGGTGGAGCCGGG..... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .................................................................................................................................................................................CAGGTGCTGTCTTCTTCTTCC.................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................AGGGAGGAGCAGCATCTGTGtat......................................................................................... | 23 | tat | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................ACTAGCTTTCTGTGACGGTGGAGCCGGG..... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ......................................................................................................................................AGGGAGGGAGGAGCAGCATC................................................................................................ | 20 | 2 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................AGGGAGGAGCAGCATCTGTG............................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................CAGGTGCTGTCTTCTTCTTCCAGa................................................. | 24 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................CAGGTGCTGTCTTCTTCTTCCA................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................TAGCTTTCTGTGACGGTGGAGCCGG...... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................ATAGAGTTCTGTTCggtt................................................................................................................................................................................................... | 18 | ggtt | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................CAGGTGCTGTCTTCTTCTTCCAGtaa............................................... | 26 | taa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ................................................................................................................................................................................................................TGACAGCTAACTAGCTTTCTGTGACGGTG............. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................AGGGAGGAGCAGCATCTGT............................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................GGGAGGGAGGAGCAGCATCTGTata.......................................................................................... | 25 | ata | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................AGGGAGGAGCAGCATCTGTGA........................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .......................................................................................................................................GGGAGGGAGGAGCAGCATCTGT............................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| GTGACACCGTTTGAAATCCTCAAAACAAGGGTGCACAATAGAGTTCTGTTCACTGAGTCCTGGTGCTTTACCTCATTCCCCAAAGTGAGCCTCCCAAGAGATGTGACTCGGGATTCCAGGAGAGCAGCTGGGGGAGGGAGGGAGGAGCAGCATCTGTGAAGGCTGCCATAGAAACCACAGGTGCTGTCTTCTTCTTCCAGGATGATTCTGACAGCTAACTAGCTTTCTGTGACGGTGGAGCCGGGGAGGA ........................................................................................................................................((((((((((((((((((((((....(((...)))....))))))))))).))))))))))).................................................... .....................................................................................................................................134...............................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT4() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................................AACCACAGGTGCTGTCTTCTTCTTCCA................................................... | 27 | 1 | 3.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - |
| ...........................................................................................................................................................................AAACCACAGGTGCTGTCTTCTTCTTCCA................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................TTCCCCAAAGTGAGCCTCCCAA......................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |