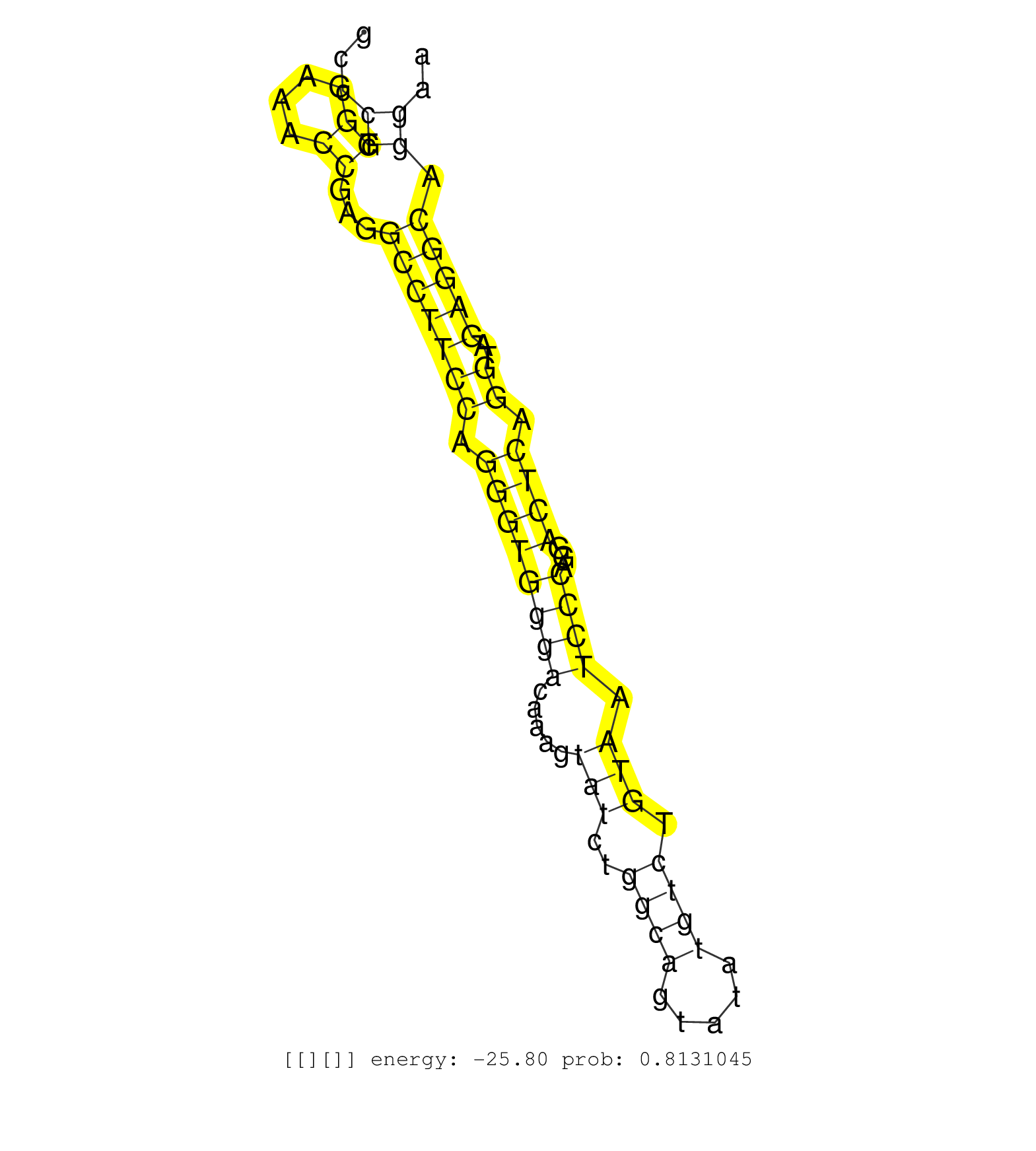

| Gene: Naglu | ID: uc007lmz.1_intron_0_0_chr11_100932542_f.5p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(13) TESTES |
| GCATCAACCTGGCTCTGGCTTGGAATGGCCAGGAGGCCATCTGGCAAAGGGTTTGTGCCAACCTGGCCCCTCACACTCTCCTGCCTGCCTTACATACACCCCTGCCCCTGGGAAACCGAGGCCTTCCAGGGTGGGACAAAGTATCTGGCAGTATATGTCTGTAATCCCAGGACTCAGGTAGAGGCAGGAAGATTCAAGATTGAGTTCCAAGGCAGGAACTCAATCAGGGCAGGTACCTGGAGGCAGGAAC ..........................................................................................................((.((....))...(((((((.((((((((.....(((..((((.....)))).))).))))...)))).))..))))).)).............................................................. .......................................................................................................104...................................................................................190.......................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................TGTAATCCCAGGACTCAGGTAGAGGCA................................................................ | 27 | 1 | 12.00 | 12.00 | 3.00 | 3.00 | 3.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | 1.00 |
| ...............................................................................................................................................................TGTAATCCCAGGACTCAGGTAGAGGC................................................................. | 26 | 1 | 9.00 | 9.00 | 3.00 | 2.00 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................TGTAATCCCAGGACTCAGGTAGAGGCAG............................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................TGGGAAACCGAGGCCTTCCAGGGTG..................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ....................................................................................................................CGAGGCCTTCCAGtc....................................................................................................................... | 15 | tc | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................................................................CACACTCTCCTGCCTGCtc................................................................................................................................................................ | 19 | tc | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................................................................................................................................................................CAGGACTCAGGTAGAGGCAGGAAGATT........................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................AGGCAGGAACTCAATCAGGGCAGGTAC.............. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................TCCCAGGACTCAGGTAGAGGCAGGAAG........................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..............................................................................................................................................................CTGTAATCCCAGGACTCAGGTAGAGG.................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .......................................................................................................................................................................CAGGACTCAGGTAGAGGCAGGAAGA.......................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...............................................................................................................................................................TGTAATCCCAGGACTCAGGTAGAGG.................................................................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| GCATCAACCTGGCTCTGGCTTGGAATGGCCAGGAGGCCATCTGGCAAAGGGTTTGTGCCAACCTGGCCCCTCACACTCTCCTGCCTGCCTTACATACACCCCTGCCCCTGGGAAACCGAGGCCTTCCAGGGTGGGACAAAGTATCTGGCAGTATATGTCTGTAATCCCAGGACTCAGGTAGAGGCAGGAAGATTCAAGATTGAGTTCCAAGGCAGGAACTCAATCAGGGCAGGTACCTGGAGGCAGGAAC ..........................................................................................................((.((....))...(((((((.((((((((.....(((..((((.....)))).))).))))...)))).))..))))).)).............................................................. .......................................................................................................104...................................................................................190.......................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|