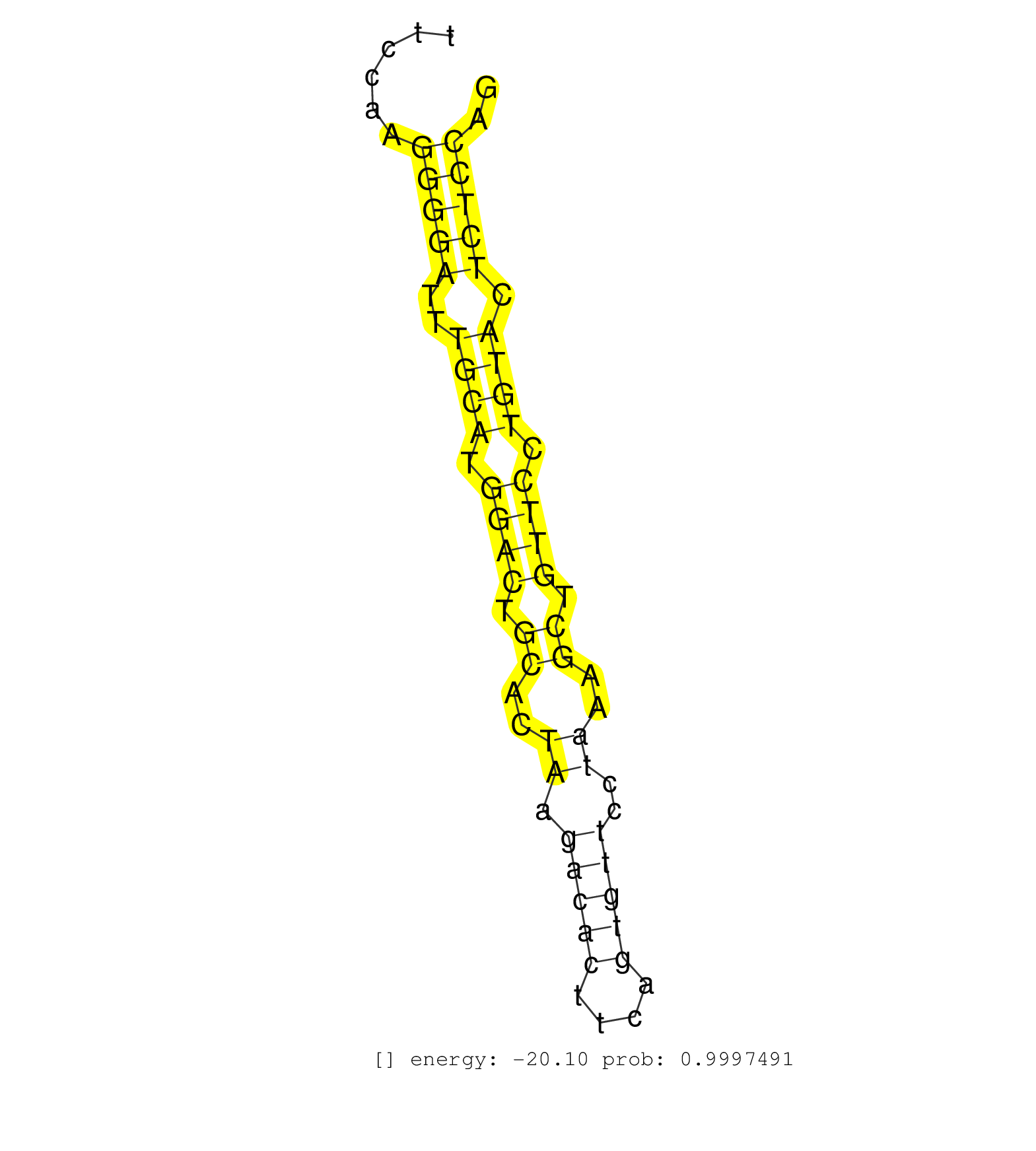

| Gene: Atp6v0a1 | ID: uc007lmt.1_intron_7_0_chr11_100890573_f.3p | SPECIES: mm9 |
 |
 |
|
(6) OTHER.mut |
(1) OVARY |
(15) TESTES |
| ATAGCTGTAGAGAATGTCAAGACTGCTCGGCATTATTGCTGTCTGCAGGCCACGTCACATGAGTGATTAATGTTCATTACCACAACAGGCCAGTCTGGCTGTGCTTATCCTGTAGTAAGCTAGGATTAGTTTCCAAGGGGATTTGCATGGACTGCACTAAGACACTTCAGTGTTCCTAAAGCTGTTCCTGTACTCTCCAGGTTCCGAGCCTCCCTCTATCCCTGCCCTGAGACTCCACAGGAAAGGAAAG ........................................................................................................................................(((((..((((.((((.((..((.(((((....)))))..))..)).)))).)))).))))).................................................... ..................................................................................................................................131..................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCCAGt................................................. | 23 | t | 14.00 | 0.00 | 1.00 | 5.00 | 4.00 | - | - | 2.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCCAGa................................................. | 23 | a | 3.00 | 0.00 | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................GGGATTTGCATGGACTGCACTAAG......................................................................................... | 24 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................AGGGGATTTGCATGGACTGCACTA........................................................................................... | 24 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCCAtt................................................. | 23 | tt | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...............GTCAAGACTGCTCGGggt......................................................................................................................................................................................................................... | 18 | ggt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................CTAAAGCTGTTCCTGTttgt....................................................... | 20 | ttgt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCCAGc................................................. | 23 | c | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................AGGGGATTTGCATGGACTGCACTAt.......................................................................................... | 25 | t | 1.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................AGGGGATTTGCATGGACTGCACTAAGAC....................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................TGTTCCTGTACTCTCCAac................................................. | 19 | ac | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCCAGttt............................................... | 25 | ttt | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................AAGGGGATTTGCATGGACTGCACTAAG......................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCCAGta................................................ | 24 | ta | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................AGGGGATTTGCATGGACTGCACTAAG......................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCCA................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ATAGCTGTAGAGAATGTCAAGACTGCTCGGCATTATTGCTGTCTGCAGGCCACGTCACATGAGTGATTAATGTTCATTACCACAACAGGCCAGTCTGGCTGTGCTTATCCTGTAGTAAGCTAGGATTAGTTTCCAAGGGGATTTGCATGGACTGCACTAAGACACTTCAGTGTTCCTAAAGCTGTTCCTGTACTCTCCAGGTTCCGAGCCTCCCTCTATCCCTGCCCTGAGACTCCACAGGAAAGGAAAG ........................................................................................................................................(((((..((((.((((.((..((.(((((....)))))..))..)).)))).)))).))))).................................................... ..................................................................................................................................131..................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................TAAGCTAGGATTAGTgccc........................................................................................................................ | 19 | gccc | 6.00 | 0.00 | 2.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ................................................................................................................TAAGCTAGGATTAGTgcc........................................................................................................................ | 18 | gcc | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................TAAGCTAGGATTAGTga........................................................................................................................ | 17 | ga | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |