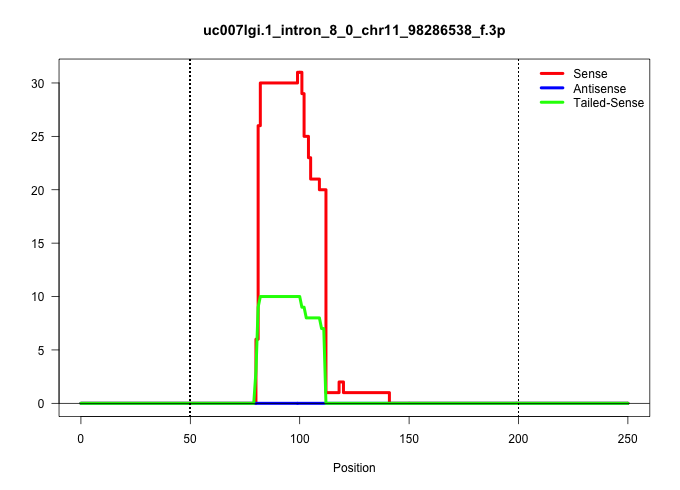
| Gene: Erbb2 | ID: uc007lgi.1_intron_8_0_chr11_98286538_f.3p | SPECIES: mm9 |
 |
 |
|
(1) PIWI.ip |
(7) TESTES |
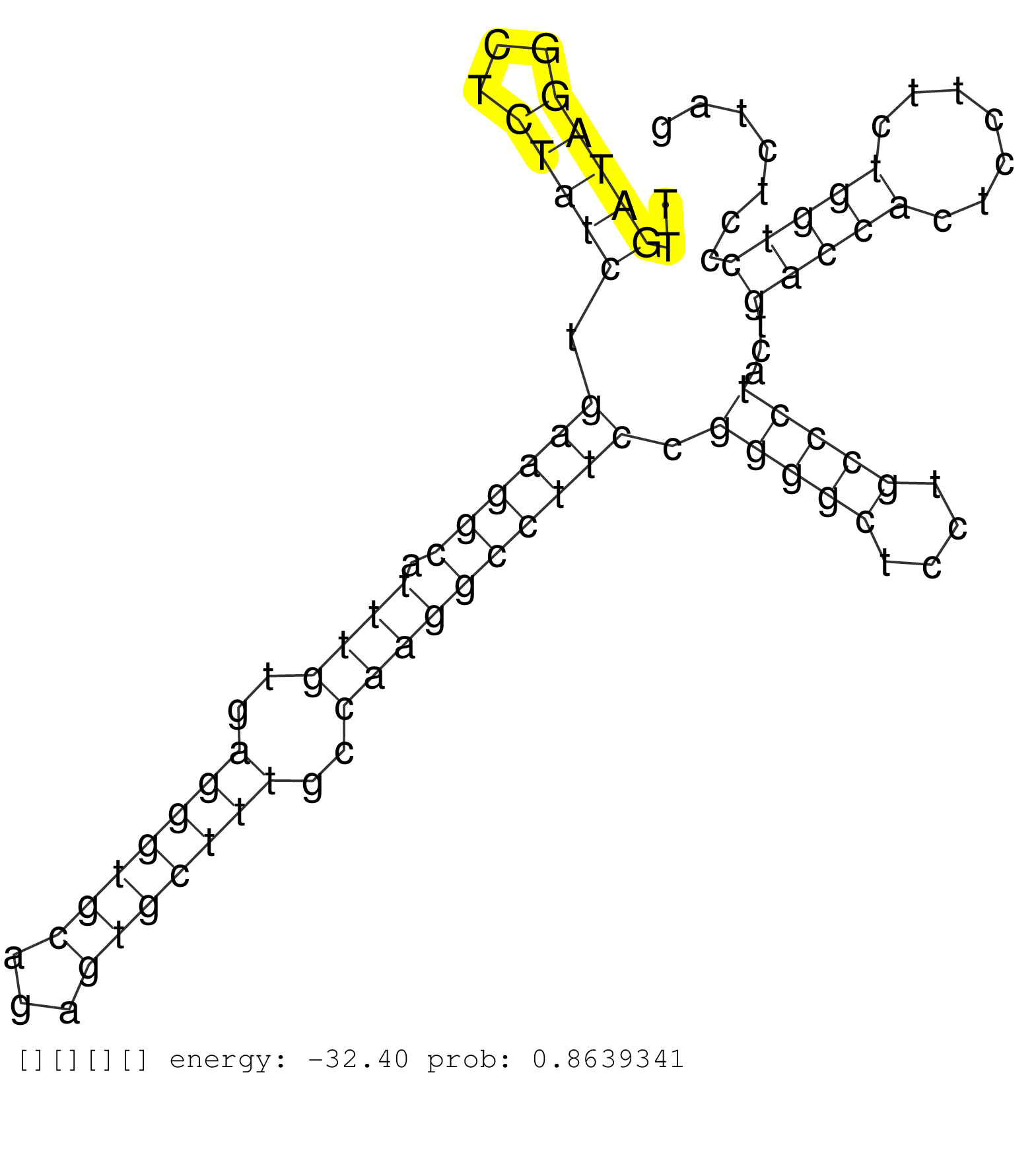
| TCTGCAGGATATACCGCAAATCAGACTTCTAGACCAAAGGACACAAATATTGCCCTGCCAGACTTTGACCCTGGGCTTTGTGCCTCAGGATGAGAACGGCTTGATAGGCTCTATCTGAAGGCATTTGTGAGGGTGCAGAGTGCTTTGCCAAGGCCTTCCGGGGCTCCTGCCCTACTGACCACTCCTTCTGGTCCCTCTAGGAACCCCTCCTCCGGCGTTGCCCCACTGAAGCCAGAGCATCTCCAAGTGT ......................................................................................................(((((...))))).((((((.((((..(((((((...)))))))..)))))))))).(((((....)))))...(((((.......)))))......................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | GSM475281(GSM475281) total RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................GCCTCAGGATGAGAACGGCTTGATAGGCTCT.......................................................................................................................................... | 31 | 1 | 14.00 | 14.00 | 4.00 | 6.00 | 4.00 | - | - | - | - |
| ................................................................................TGCCTCAGGATGAGAACGGCTTGATAGGCTCT.......................................................................................................................................... | 32 | 1 | 5.00 | 5.00 | - | 3.00 | 2.00 | - | - | - | - |
| .................................................................................GCCTCAGGATGAGAACGGCTTGATAGGgtct.......................................................................................................................................... | 31 | gtct | 3.00 | 0.00 | 3.00 | - | - | - | - | - | - |
| .................................................................................GCCTCAGGATGAGAACGGCTT.................................................................................................................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - |
| .................................................................................GCCTCAGGATGAGAACGGCTTGATAGGC............................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - |
| ................................................................................TGCCTCAGGATGAGAACGGCTT.................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - |
| .................................................................................GCCTCAGGATGAGAACGGCTTGATAGGCTCc.......................................................................................................................................... | 31 | c | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - |
| .................................................................................GCCTCAGGATGAGAACGGCTTGAT................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - |
| ..................................................................................CCTCAGGATGAGAACGGCTT.................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - |
| .................................................................................GCCTCAGGATGAGAACGGCTTt................................................................................................................................................... | 22 | t | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - |
| ...................................................................................................CTTGATAGGCTCTATCTGAAG.................................................................................................................................. | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - |
| ..................................................................................CCTCAGGATGAGAACGGCTTGA.................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - |
| ..................................................................................CCTCAGGATGAGAACGGCT..................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - |
| ................................................................................TGCCTCAGGATGAGAACGGCTTGATAGGCTCc.......................................................................................................................................... | 32 | c | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - |
| ......................................................................................................................AGGCATTTGTGAGGGTGCAGAGT............................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - |
| .................................................................................GCCTCAGGATGAGAACGGCa..................................................................................................................................................... | 20 | a | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - |
| ..................................................................................CCTCAGGATGAGAACGGCTTGATAGGgt............................................................................................................................................ | 28 | gt | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - |
| .................................................................................GCCTCAGGATGAGAACGGCTTGA.................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - |
| ................................................................................TGCCTCAGGATGAGAACGGCTTGATAGGgtct.......................................................................................................................................... | 32 | gtct | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - |
| .................................................................................GCCTCAGGATGAGAACGGCT..................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - |
| ................................................................................TGCCTCAGGATGAGAACGGCTTGATAGGgttt.......................................................................................................................................... | 32 | gttt | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - |
| ........................................................................................................TAGGCTCTATCTGAAGGCA............................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 |
| ..................................................................................CCTCAGGATGAGAACGGCTTGAT................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - |
| TCTGCAGGATATACCGCAAATCAGACTTCTAGACCAAAGGACACAAATATTGCCCTGCCAGACTTTGACCCTGGGCTTTGTGCCTCAGGATGAGAACGGCTTGATAGGCTCTATCTGAAGGCATTTGTGAGGGTGCAGAGTGCTTTGCCAAGGCCTTCCGGGGCTCCTGCCCTACTGACCACTCCTTCTGGTCCCTCTAGGAACCCCTCCTCCGGCGTTGCCCCACTGAAGCCAGAGCATCTCCAAGTGT ......................................................................................................(((((...))))).((((((.((((..(((((((...)))))))..)))))))))).(((((....)))))...(((((.......)))))......................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | GSM475281(GSM475281) total RNA. (testes) |
|---|