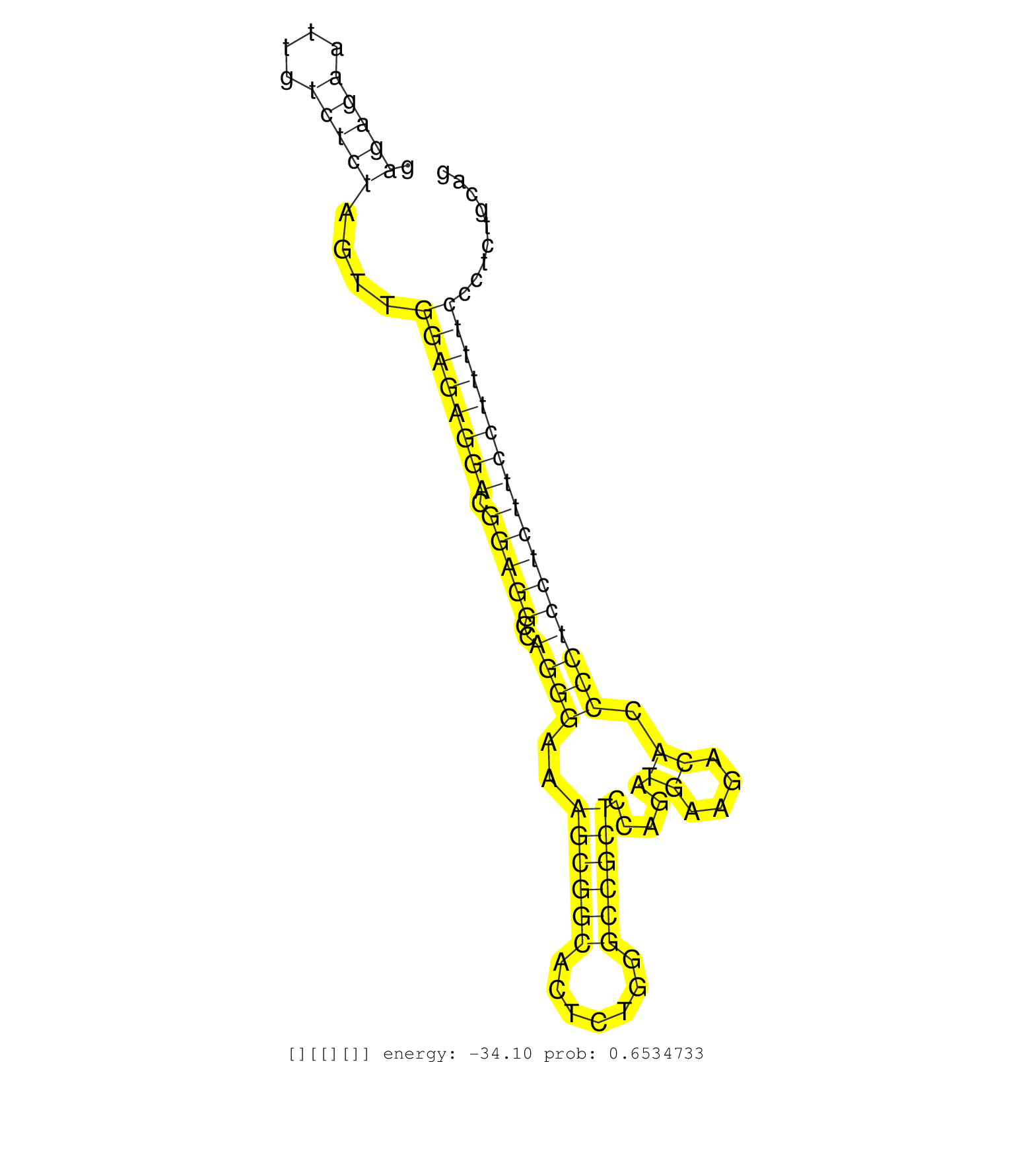

| Gene: 4933428G20Rik | ID: uc007lec.1_intron_7_0_chr11_97326171_f | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(2) PIWI.ip |
(1) TDRD1.ip |
(11) TESTES |
| GCCAACCAAGCTCTGATCAGCAAGAAGCTTAATGACTATCGCAAAGTGAGGTAAGGCCAGTTCTCCCAGAGCAGCCTGTGGTGGGGAGGAAAGCCTATGGGGTTGGGAGTTTCCAGGCAGAGATCTCTAGTCCTCGCTCAGACAAGTGAGCCGAGAGAATTGTCTCTAGTTGGAGAGGACGGAGGCCAGGGAAAGCGGCACTCTGGGCCGCTCCAGATGAAGACACCCCTCCTCTTCCTTTTCCCTCTGCAGCCATAGCTCTGGGCCCAAAGCCGATTCCTCCCCTAAAGGCTCTCGGGGCC .........................................................................................................................................................(((((....)))))....((((((((.(((((..((((..((((((.......)))))).....((....)).)))))))))))))))))........................................................... ........................................................................................................................................................153................................................................................................252................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM475281(GSM475281) total RNA. (testes) | mjTestesWT2() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................AGTTGGAGAGGACGGAGGCCAGGGAAAGCGGCACTCTGGGCCGCTCCAGATG................................................................................... | 52 | 1 | 50.00 | 50.00 | 50.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................................TAGCTCTGGGCCCAAAGCCGAT......................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...........TCTGATCAGCAAGAAGCTTAATGACT......................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................AAGAAGCTTAATGACTATCGCAAAGTG.............................................................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................................................................................................................................................AGAGAATTGTCTCTAGTTGGAGAGGACGGA....................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................AGCAAGAAGCTTAATGACTATCGC.................................................................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...........................................................................................................................................................................GGAGAGGACGGAGGCCAGGGAAAG........................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......AAGCTCTGATCAGCAAGAAGCT................................................................................................................................................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........................GCTTAATGACTATCGCAAAGTGAGc........................................................................................................................................................................................................................................................... | 25 | c | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .............TGATCAGCAAGAAGCTTAATGACTATC...................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...............ATCAGCAAGAAGCTTAATGACTATCG..................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............TGATCAGCAAGAAGCTTAATGACTATCGC.................................................................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| GCCAACCAAGCTCTGATCAGCAAGAAGCTTAATGACTATCGCAAAGTGAGGTAAGGCCAGTTCTCCCAGAGCAGCCTGTGGTGGGGAGGAAAGCCTATGGGGTTGGGAGTTTCCAGGCAGAGATCTCTAGTCCTCGCTCAGACAAGTGAGCCGAGAGAATTGTCTCTAGTTGGAGAGGACGGAGGCCAGGGAAAGCGGCACTCTGGGCCGCTCCAGATGAAGACACCCCTCCTCTTCCTTTTCCCTCTGCAGCCATAGCTCTGGGCCCAAAGCCGATTCCTCCCCTAAAGGCTCTCGGGGCC .........................................................................................................................................................(((((....)))))....((((((((.(((((..((((..((((((.......)))))).....((....)).)))))))))))))))))........................................................... ........................................................................................................................................................153................................................................................................252................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM475281(GSM475281) total RNA. (testes) | mjTestesWT2() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................................................................ACACCCCTCCTCTTCCtta................................................................ | 19 | tta | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................CTCTGGGCCGCTCCAGga...................................................................................... | 18 | ga | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |