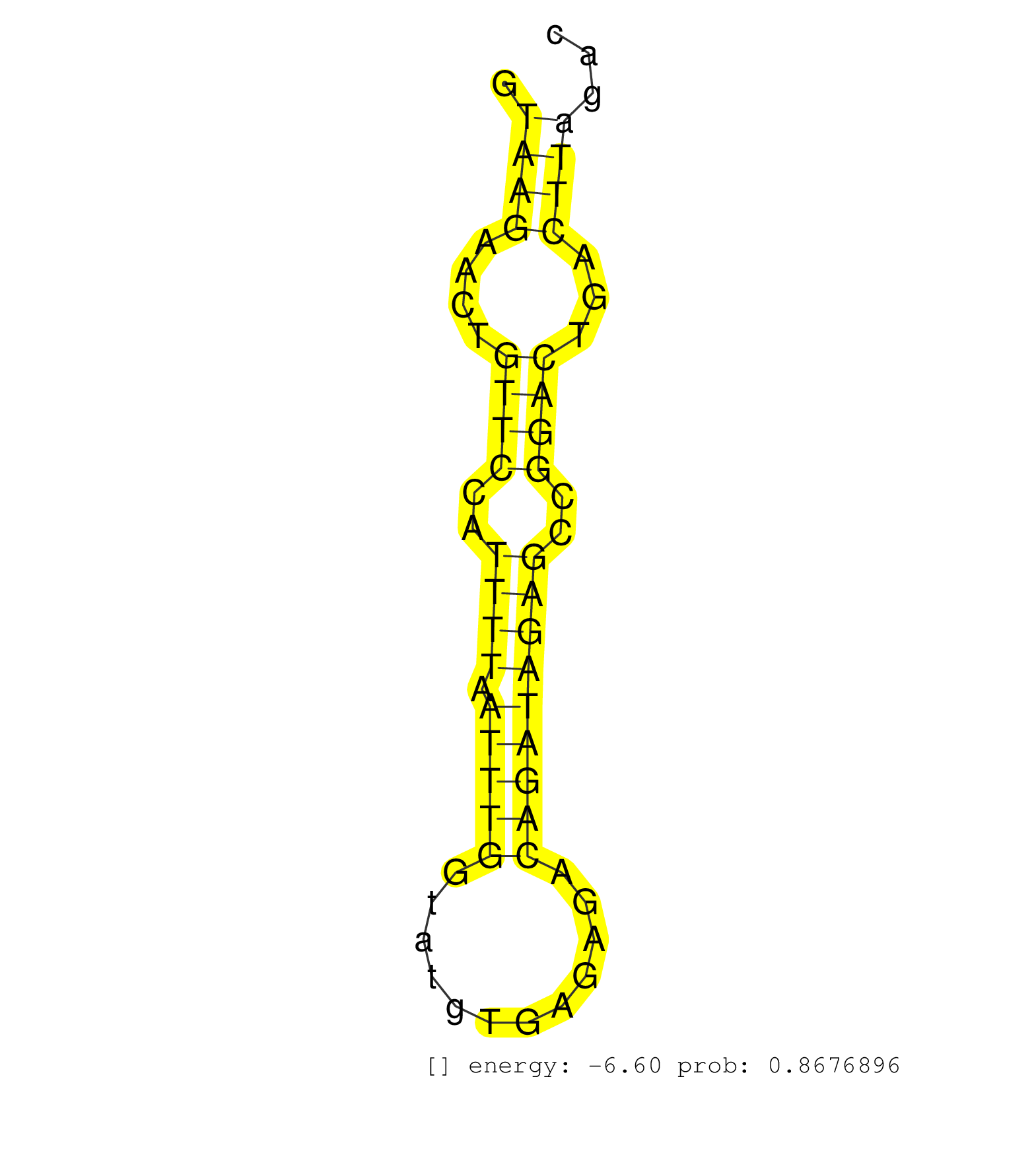

| Gene: Npepps | ID: uc007ldv.1_intron_20_0_chr11_97119666_r.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(2) PIWI.ip |
(13) TESTES |
| TGCTGATATTGACATTATTACAGCTTCATATGCCCCAGAAGGAGATGAAGGTAAGAACTGTTCCATTTTAATTTGGTATGTGAGAGACAGATAGAGCCGGACTGACTTAGACTTTCAGTCTGAAACTTCTATACTGCCTGCTTTAAAATGTGCTGGGCACGCTGATACATTCCTGTAATCCCACGATTTGTGAGGCTGAGGCAAGAGGACTGTATTCTTCTTAAAAAGACCTCACCACCTTGAACCTTGA ...................................................((((....((((..((((.(((((............)))))))))..))))...))))............................................................................................................................................. ..................................................51...........................................................112........................................................................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT4() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGAACTGTTCCATTTTAATTTGG.............................................................................................................................................................................. | 26 | 1 | 10.00 | 10.00 | 2.00 | 4.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGAACTGTTCCATTTTA.................................................................................................................................................................................... | 20 | 1 | 5.00 | 5.00 | 1.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................TGAGAGACAGATAGAGCCGGACTGACTT.............................................................................................................................................. | 28 | 1 | 3.00 | 3.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ................................................................................TGAGAGACAGATAGAGCCGGACTGACT............................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................AAGAACTGTTCCATTTTAATTTGGTAT........................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TATACTGCCTGCTTTAAAATGTGCTG............................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................ACAGCTTCATATGCCCCAGAAGGAGA............................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................GTAAGAACTGTTCCATTTTAATT................................................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGAACTGTTCCATTTTAATTTGGT............................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TAATCCCACGATTTGTGAGGCTGAGGCA............................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................................................AGAGACAGATAGAGCCGGACT................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ........................................................................................................................................................................................................GCAAGAGGACTGTATTCTTCTTAAAAAGACC................... | 31 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................TGTGAGAGACAGATAGAGCCGGACTG.................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TTCTATACTGCCTGCTTTAAAATGTG.................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................TAGAGCCGGACTGACTTAGACTTTCA..................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ....................................................................................................................................................................................CCACGATTTGTGAGGCTGAGGCAAGAGG.......................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................TGGTATGTGAGAGACAGATAGAGCCGG...................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .................................CCCAGAAGGAGATGAAGGTAAGA.................................................................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TCCCACGATTTGTGAGGCTGAGGCAA.............................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................................................................................................................................................................................CGATTTGTGAGGCTGAGGCAAGAGGA......................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............TTATTACAGCTTCATATGCCCCAGAA.................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................GTAAGAACTGTTCCATTTTAATTTGGa............................................................................................................................................................................. | 27 | a | 1.00 | 10.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................TTTGTGAGGCTGAGGCAAGAagac........................................ | 24 | agac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................GTAAGAACTGTTCCATTTTAATTTGGTA............................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................TTGTGAGGCTGAGGCAAGAGG.......................................... | 21 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - |
| TGCTGATATTGACATTATTACAGCTTCATATGCCCCAGAAGGAGATGAAGGTAAGAACTGTTCCATTTTAATTTGGTATGTGAGAGACAGATAGAGCCGGACTGACTTAGACTTTCAGTCTGAAACTTCTATACTGCCTGCTTTAAAATGTGCTGGGCACGCTGATACATTCCTGTAATCCCACGATTTGTGAGGCTGAGGCAAGAGGACTGTATTCTTCTTAAAAAGACCTCACCACCTTGAACCTTGA ...................................................((((....((((..((((.(((((............)))))))))..))))...))))............................................................................................................................................. ..................................................51...........................................................112........................................................................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT4() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|