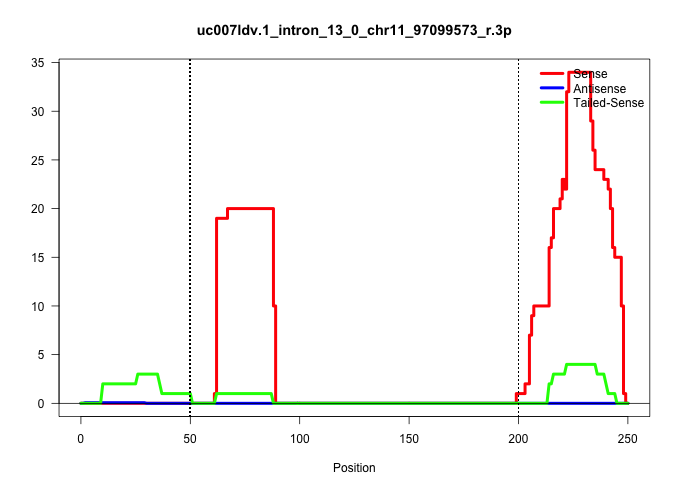
| Gene: Npepps | ID: uc007ldv.1_intron_13_0_chr11_97099573_r.3p | SPECIES: mm9 |
 |
 |
|
(8) OTHER.mut |
(5) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(26) TESTES |
| AGCAGTCACATGGTGGCTCACAACCATCTGTCATGGGATCTGATGCCCTCAAATATGAGTTCATTACTAACTGGTTTCTGTTGAGTTTTACTTTACAAAATTACAAATTTTCAAGTACAATAGAAGGGAGACATGTTTTTCCAGTAATGCAGTGACTATTCCTATGTGTCCTTCACTCGATCTGCCTTTTTCTTTTTTAGGAATGGTGGACTCATCTCTGGTTGAATGAAGGCTTTGCATCCTGGATTGA ........................................................................................................................(((((((((((...((...((.((...((.((.((((((....))).))))).)))).))...))...)))))))))))................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT1() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................ATTACTAACTGGTTTCTGTTGAGTTT.................................................................................................................................................................. | 26 | 1 | 10.00 | 10.00 | - | 5.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | 1.00 | - | - |
| ..............................................................ATTACTAACTGGTTTCTGTTGAGTTTT................................................................................................................................................................. | 27 | 1 | 9.00 | 9.00 | - | 1.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | 1.00 | 1.00 | 1.00 | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - |
| ..............................................................................................................................................................................................................................TGAATGAAGGCTTTGCATCCTGGATT.. | 26 | 1 | 8.00 | 8.00 | 2.00 | - | - | 3.00 | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................GAATGGTGGACTCATCTCTGGTTG.......................... | 24 | 1 | 5.00 | 5.00 | - | - | - | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................GTGGACTCATCTCTGGTTGAATGAAGGC................. | 28 | 1 | 3.00 | 3.00 | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TGAATGAAGGCTTTGCATCCTGGAT... | 25 | 1 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................TCTCTGGTTGAATGAAGGCTTTGCATCCT....... | 29 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................TCTCTGGTTGAATGAAGGCTTTGCATCC........ | 28 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................GTGGACTCATCTCTGGTTGAATGAAGGCT................ | 29 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................GTTGAATGAAGGCTTTGCATCCTGGAT... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..............................................................ATTACTAACTGGTTTCTGTTGAGgtt.................................................................................................................................................................. | 26 | gtt | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................GGACTCATCTCTGGTTGAATGAAGGCTT............... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TGGACTCATCTCTGGTTGAATGAAGGCTT............... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................GTGGACTCATCTCTGGTTGAATGA..................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................CTCTGGTTGAATGAAGGCTTTGCATCCT....... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TGAATGAAGGCTTTGCAa.......... | 18 | a | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................TGGTGGACTCATCTCTGGTTGAATGAAGGC................. | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TCTGGTTGAATGAAGGCTTTGCATCCT....... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................TGGTGGACTCATCTCTGGTTGAATGA..................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................TCTGTCATGGGATCTGATGCCCctg....................................................................................................................................................................................................... | 25 | ctg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TGGACTCATCTCTGGTTGAATGAAGG.................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................TCTCTGGTTGAATGAAGGCTTa.............. | 22 | a | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................GAATGAAGGCTTTGCATCCTGGATTG. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................TACTAACTGGTTTCTGTTGA...................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................GGAATGGTGGACTCATCTCTGG............................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................TCTCTGGTTGAATGAAGGCTTTGCATC......... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TCTGGTTGAATGAAGGCTTTGCA........... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................TAACTGGTTTCTGTTGAGTTT.................................................................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................TCTCTGGTTGAATGAAGGCT................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................GGTTGAATGAAGGCTTTGCATCCTGGAT... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................TCTCTGGTTGAATGAAGGCTTTGaatc......... | 27 | aatc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .............................................................CATTACTAACTGGTTTCTGTTGAGTTTT................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TCTGGTTGAATGAAGGCTTTGCATCCTaa..... | 29 | aa | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................GAATGAAGGCTTTGCATCCTGGATT.. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TGGACTCATCTCTGGTTGAATGAAGGC................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........TGGTGGCTCACAACCATCTGTCATGta..................................................................................................................................................................................................................... | 27 | ta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........TGGTGGCTCACAACCATCTGTCgtgt...................................................................................................................................................................................................................... | 26 | gtgt | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TCTGGTTGAATGAAGGCTTTGCATCCTG...... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGCAGTCACATGGTGGCTCACAACCATCTGTCATGGGATCTGATGCCCTCAAATATGAGTTCATTACTAACTGGTTTCTGTTGAGTTTTACTTTACAAAATTACAAATTTTCAAGTACAATAGAAGGGAGACATGTTTTTCCAGTAATGCAGTGACTATTCCTATGTGTCCTTCACTCGATCTGCCTTTTTCTTTTTTAGGAATGGTGGACTCATCTCTGGTTGAATGAAGGCTTTGCATCCTGGATTGA ........................................................................................................................(((((((((((...((...((.((...((.((.((((((....))).))))).)))).))...))...)))))))))))................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT1() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..CAGTCACATGGTGGCTCACAACCATCTG............................................................................................................................................................................................................................ | 28 | 16 | 0.06 | 0.06 | - | 0.06 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |