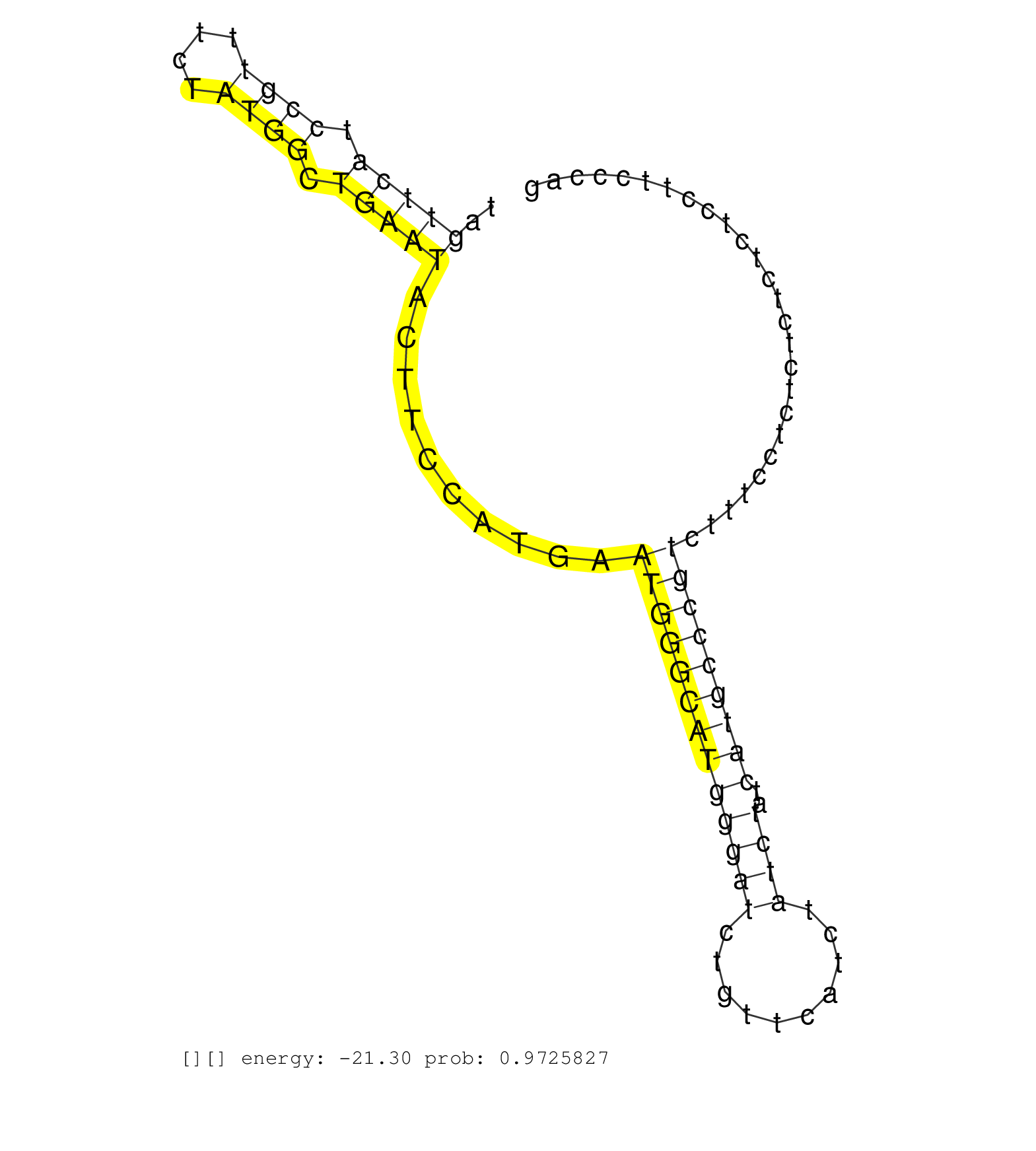

| Gene: Snx11 | ID: uc007lci.1_intron_0_0_chr11_96630637_r.3p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(5) PIWI.ip |
(1) PIWI.mut |
(17) TESTES |
| CTCTCCTGCCATGGGTCCTGGGATTGAAGTCAGTCATCAGGCTTGGCAGCGGGGGGCCTTCCACCCACTGAGCCACTCTGTCGGCCCCAGCGTGTCTCCGTAGTTCATCCGTTTCTATGGCTGAATACTTCCATGAATGGGCATGGGATCTGTTCATCTATCTATCATGCCCGTCTTTCCTCTCTCTCTCTCCTTCCCAGTTGCTGCTTTCTTCCAAGATCAGGGCGCAGGAGCTCCCCTTCGCCGCCTC ......................................................................................................(((((.((((....)))).)))))..........(((((((((((((..........))))..)))))))))............................................................................ ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesWT2() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................TATGGCTGAATACTTCCATGAATGGGCAT.......................................................................................................... | 29 | 1 | 12.00 | 12.00 | 3.00 | 3.00 | 4.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - |
| ...............................................................................................................................................................................................................TTTCTTCCAAGATCAGGGCGCAGGAGC................ | 27 | 1 | 4.00 | 4.00 | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................CCCAGTTGCTGCTTTCTTCCAAGATCA............................ | 27 | 1 | 3.00 | 3.00 | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TATGGCTGAATACTTCCATGAATGGGC............................................................................................................ | 27 | 1 | 3.00 | 3.00 | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................TGGCTGAATACTTCCATGAATGGGCAT.......................................................................................................... | 27 | 1 | 3.00 | 3.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................CCAAGATCAGGGCGCAGGAGCTCCCCTT......... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................................TGGCTGAATACTTCCATGAATGGGCATG......................................................................................................... | 28 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TATGGCTGAATACTTCCATGAATGGGCA........................................................................................................... | 28 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TCCTGCCATGGGTCCTGGGATTGAA.............................................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................TTCCAAGATCAGGGCGCAGGAGCTCCC............ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................TTCCAAGATCAGGGCGCAGGAGCTCCCCT.......... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ....................................................................................................TAGTTCATCCGTTTCTATGGCTGAAT............................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......TGCCATGGGTCCTGGGATTGAAGTC........................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ................................................................................................................TTCTATGGCTGAATACTTCCATGAATGGGC............................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TATGGCTGAATACTTCCATGAATGGG............................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CTCTCCTGCCATGGGTCCTGGGATTGAAGTCAGTCATCAGGCTTGGCAGCGGGGGGCCTTCCACCCACTGAGCCACTCTGTCGGCCCCAGCGTGTCTCCGTAGTTCATCCGTTTCTATGGCTGAATACTTCCATGAATGGGCATGGGATCTGTTCATCTATCTATCATGCCCGTCTTTCCTCTCTCTCTCTCCTTCCCAGTTGCTGCTTTCTTCCAAGATCAGGGCGCAGGAGCTCCCCTTCGCCGCCTC ......................................................................................................(((((.((((....)))).)))))..........(((((((((((((..........))))..)))))))))............................................................................ ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesWT2() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................GCGTGTCTCCGTAGTTCATCCGTTTCTA..................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................................................GTGTCTCCGTAGTTCATCCGTTTCTA..................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................GTTGCTGCTTTCTTCCAattt.................................. | 21 | attt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .........................................................................................GCGTGTCTCCGTAGTTCATCCGTT......................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...................................................................................................................................................................................................CCCAGTTGCTGCTTTCTTCCA.................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................CTCCGTAGTTCAgcat............................................................................................................................................... | 16 | gcat | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................CATCCGTTTCTATGGCTGAATACTTCCA..................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................CGTGTCTCCGTAGTTCATCCGTTTCTA..................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ....................................................................................................................................................................................................CAGTTGCTGCTTTCTTCCc................................... | 19 | c | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |