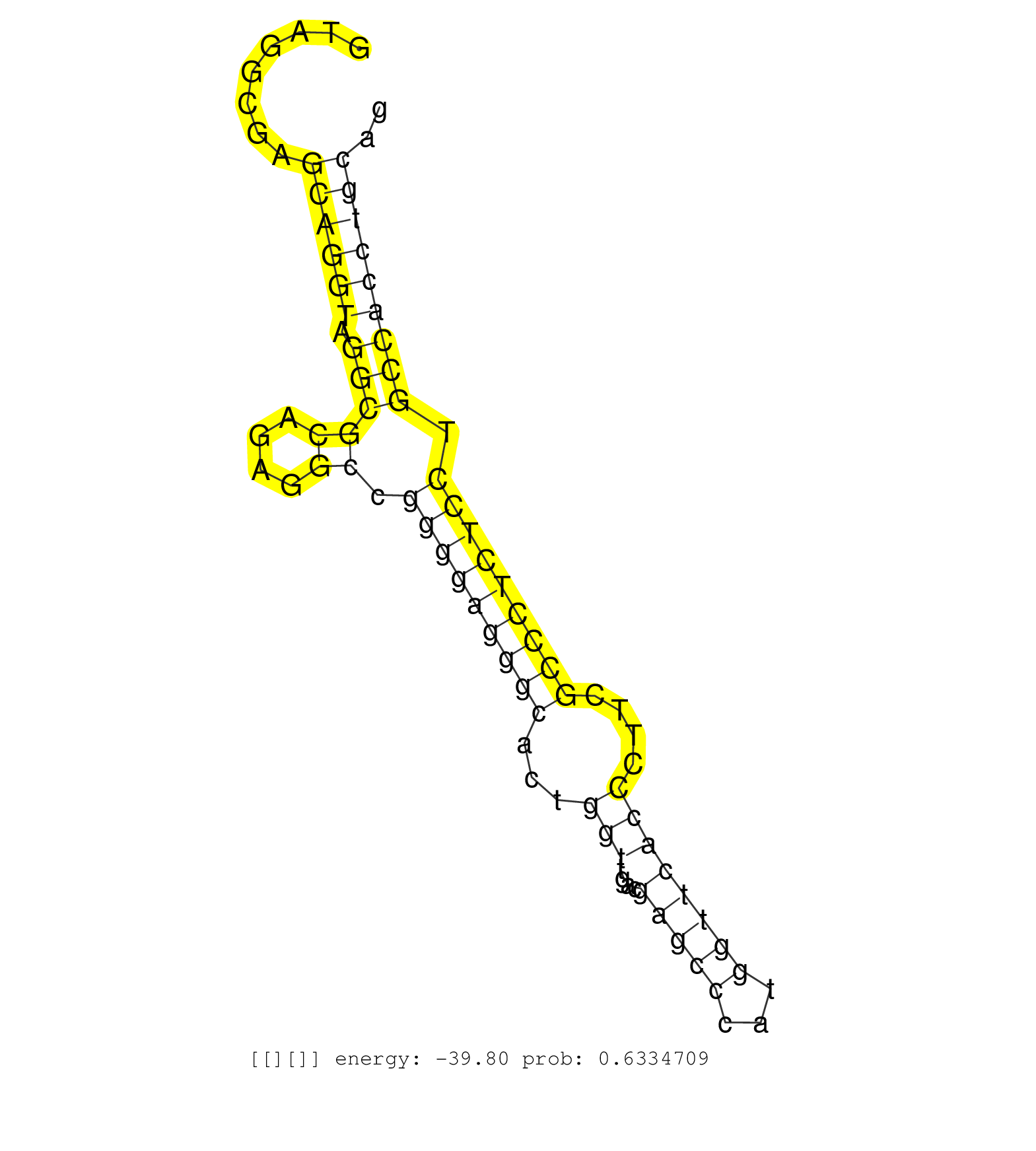
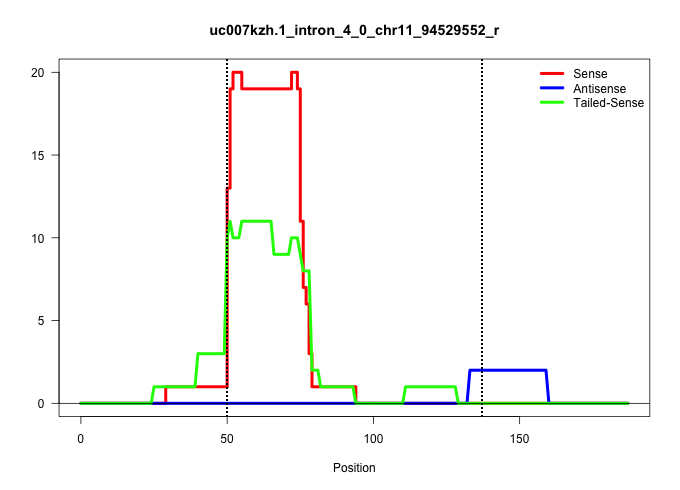
| Gene: Xylt2 | ID: uc007kzh.1_intron_4_0_chr11_94529552_r | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(5) PIWI.ip |
(1) TDRD1.ip |
(20) TESTES |
| TCGTGGCCCAGCTTCGCCAGTTCTATACATACACATTGCTTCCGGCTGAGGTAGGCGAGCAGGTAGGCGCAGAGGCCGGGGAGGGCACTGGTTGACGAGCCCATGGTTCACCCTTCGCCCTCTCCTGCCACCTGCAGTCATTCTTCCACACAGTGCTGGAGAACAGCCCGGCCTGTGCGAGCCTGGT ..........................................................((((((.(((((....)).(((((((((...(((....(((((...))))))))....))))))))).))))))))).................................................... ..................................................51....................................................................................137................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | mjTestesWT1() Testes Data. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAGGCGAGCAGGTAGGCGCAGAGG................................................................................................................ | 25 | 1 | 7.00 | 7.00 | 2.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................GTAGGCGAGCAGGTAGGCGCAGAGGCCGt............................................................................................................ | 29 | t | 4.00 | 0.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGCGAGCAGGTAGGCGCAGAGGC............................................................................................................... | 26 | 1 | 3.00 | 3.00 | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................TCCGGCTGAGGTAGGCGAGCAGtgct......................................................................................................................... | 26 | tgct | 2.00 | 0.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAGGCGAGCAGGTAGGCGCAGAGGCCG............................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - |
| .....................................................................................................................................................ACAGTGCTGGAGAACAG..................... | 17 | 2 | 1.50 | 1.50 | - | - | - | - | - | - | - | - | - | 1.50 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................AGGCCGGGGAGGGCACTGGTTa............................................................................................. | 22 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...................................................TAGGCGAGCAGGTAGGCGCAGAGGCCGt............................................................................................................ | 28 | t | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGCGAGCAGGTAGGCGCAGAGGCCGG............................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................AGGCCGGGGAGGGCACTGGTTG............................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGCGAGCAGGTAGGCGCAGAGt................................................................................................................ | 25 | t | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................TACATACACATTGCTTCCGGCTGAGtc....................................................................................................................................... | 27 | tc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...................................................TAGGCGAGCAGGTAGGCGCAGAGG................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .......................................................CGAGCAGGTAGGCGCAGAGGCCGGGag......................................................................................................... | 27 | ag | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................CCTTCGCCCTCTCCcctg.......................................................... | 18 | cctg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ....................................................AGGCGAGCAGGTAGGCGCAGAGGCCG............................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGCGAGCAGGTAGGCGCAGAGGCCtt............................................................................................................ | 29 | tt | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAGGCGAGCAGGTAGGCGCAGAGGC............................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGCGAGCAGGTAGGCGCAGAGta............................................................................................................... | 26 | ta | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAGGCGAGCAGGTAGGCGCAGAGGCC.............................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGCGAGCAGGTAGGCGCAGAG................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................TACACATTGCTTCCGGCTGAGGTAGG.................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAGGCGAGCAGGTAGGCGCAGAGGCCGG............................................................................................................ | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TCGTGGCCCAGCTTCGCCAGTTCTATACATACACATTGCTTCCGGCTGAGGTAGGCGAGCAGGTAGGCGCAGAGGCCGGGGAGGGCACTGGTTGACGAGCCCATGGTTCACCCTTCGCCCTCTCCTGCCACCTGCAGTCATTCTTCCACACAGTGCTGGAGAACAGCCCGGCCTGTGCGAGCCTGGT ..........................................................((((((.(((((....)).(((((((((...(((....(((((...))))))))....))))))))).))))))))).................................................... ..................................................51....................................................................................137................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | mjTestesWT1() Testes Data. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................GCAGTCATTCTTCCACACAGTGCTGGA........................... | 27 | 1 | 3.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 |
| ......................................................................................ACTGGTTGACGAGCCCATGGTTCA............................................................................. | 24 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |