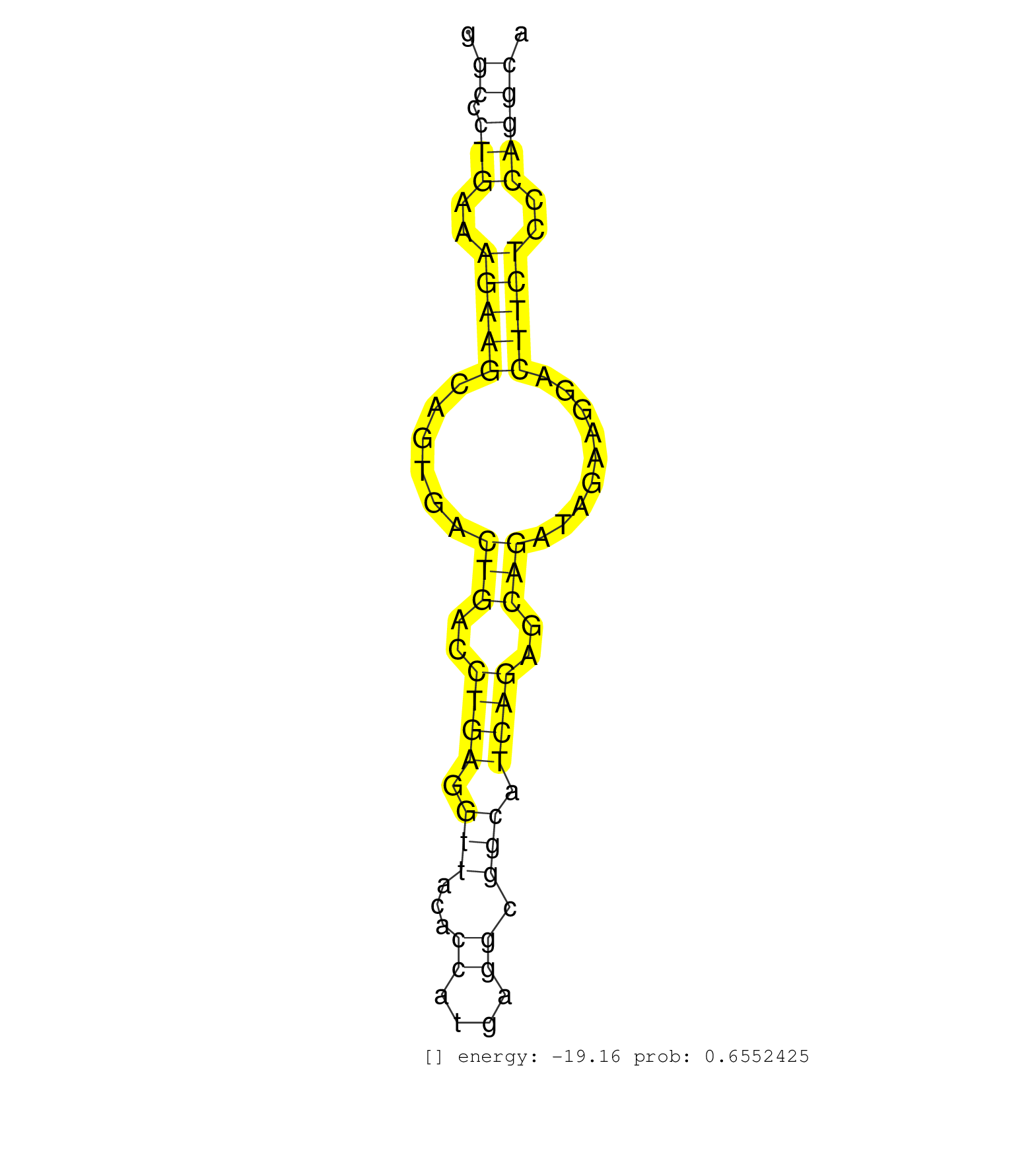
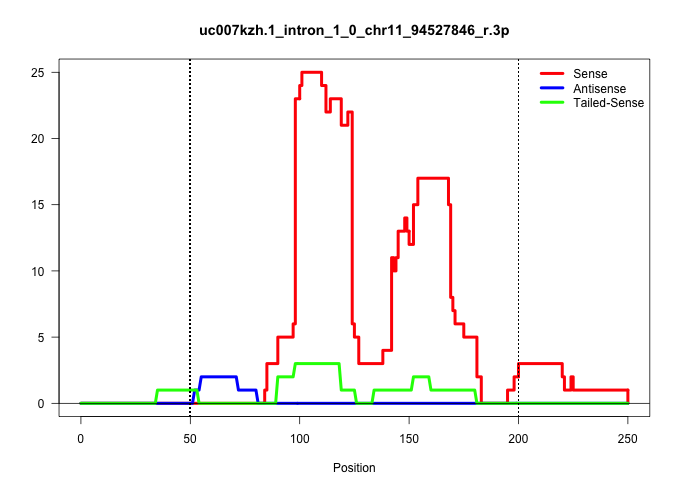
| Gene: Xylt2 | ID: uc007kzh.1_intron_1_0_chr11_94527846_r.3p | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(1) OVARY |
(5) PIWI.ip |
(2) PIWI.mut |
(1) TDRD1.ip |
(21) TESTES |
| ACAGTTCAGGTGGCAGCCTGGAATAAACCTCAGAGCTGAGAGAGATGTTTCAGTTCTAGACCTACCCCTTCAATTTGAAGATAGGGAAACTGAGGCCCTGAAAGAAGCAGTGACTGACCTGAGGTTACACCATGAGGCGGCATCAGAGCAGATAGAAGGACTTCTCCCAGGCATCTGCCCAGCTGCCTCCCTCCCTACAGGTTGGTACTGAGTGGGACCCCAAAGAACGTCTCTTCCGGAACTTTGGGGG ..............................................................................................((.(((..(((((......(((..((((.(((...((....)).))).))))..))).........)))))..))))).............................................................................. .............................................................................................94.............................................................................173........................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................TGAAAGAAGCAGTGACTGACCTGAGG.............................................................................................................................. | 26 | 1 | 15.00 | 15.00 | 4.00 | 4.00 | 1.00 | 3.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TCAGAGCAGATAGAAGGACTTCTCCCA................................................................................. | 27 | 1 | 6.00 | 6.00 | 1.00 | 1.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................TAGAAGGACTTCTCCCAGGCATCTGCCCA..................................................................... | 29 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................TAGGGAAACTGAGGCCCTGAAAGAAG............................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................GAAGGACTTCTCCCAGGCATCTGCCCAGC................................................................... | 29 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................GAAACTGAGGCCCTGAAAGAAGCAGTG.......................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................TGAAAGAAGCAGTGACTGACCTGAGGT............................................................................................................................. | 27 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................TGAGGCCCTGAAAGAAGCAGTGACTGACC................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TGACCTGAGGTTACACCATGAGGCGGCAT........................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................AGGCGGCATCAGAGCAGATAGAAGGt.......................................................................................... | 26 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................AAGGACTTCTCCCAGGCATCTGCCCAGC................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ........................................................................................................................................................................................................GTTGGTACTGAGTGGGACCCCAAAG......................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TTGGTACTGAGTGGGACCCCAAAa......................... | 24 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................CTGAGAGAGATGTTTgatt.................................................................................................................................................................................................... | 19 | gatt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................GAGCAGATAGAAGGACTTCTCCCAG................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TACTGAGTGGGACCCCAAAGAACGacac................. | 28 | acac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...................................................................................................................................................................................................TACAGGTTGGTACTGAGTGGGACCCC............................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................TACACCATGAGGCGGCATCAGAGCA.................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................................................................TGAAAGAAGCAGTGACTGACCTGAGGat............................................................................................................................ | 28 | at | 1.00 | 15.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................TGAGGCCCTGAAAGAAGCAGTGACTaacc................................................................................................................................... | 29 | aacc | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................GGCATCAGAGCAGATAGAAGGACTTCTCCC.................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................TAGAAGGACTTCTCCCAGGCATCTGacct..................................................................... | 29 | acct | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................AGAGCAGATAGAAGGACTTCTCCCA................................................................................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................GGAAACTGAGGCCCTGAAAGAAGCAG............................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................TGAGGCCCTGAAAGAAGCAGTGACTGACt................................................................................................................................... | 29 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................GAGCAGATAGAAGGACTTCTCCCAGG............................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................GTTGGTACTGAGTGGGACCCCAAAGA........................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................AAAGAAGCAGTGACTGACCTGAGGTTA........................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................CTGAAAGAAGCAGTGACTGACCTGAGG.............................................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................GAACGTCTCTTCCGGAACTTTGGGGG | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ......................................................................................................................................................................................................AGGTTGGTACTGAGTGGGACCC.............................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................CAGATAGAAGGACTTCTCCCAGGCATC........................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................AAGAAGCAGTGACTGACCTGAGGTTA........................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................................................................................GGTTACACCATGAGGCGGCATCAGAGC..................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TCAGAGCAGATAGAAGGACTTCTCCC.................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ACAGTTCAGGTGGCAGCCTGGAATAAACCTCAGAGCTGAGAGAGATGTTTCAGTTCTAGACCTACCCCTTCAATTTGAAGATAGGGAAACTGAGGCCCTGAAAGAAGCAGTGACTGACCTGAGGTTACACCATGAGGCGGCATCAGAGCAGATAGAAGGACTTCTCCCAGGCATCTGCCCAGCTGCCTCCCTCCCTACAGGTTGGTACTGAGTGGGACCCCAAAGAACGTCTCTTCCGGAACTTTGGGGG ..............................................................................................((.(((..(((((......(((..((((.(((...((....)).))).))))..))).........)))))..))))).............................................................................. .............................................................................................94.............................................................................173........................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................GTTCTAGACCTACCCCTTCA.................................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................................GCGGCATCAGAGCAgaga.................................................................................................... | 18 | gaga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .......................ACCTCAGAGCTGAGAagt................................................................................................................................................................................................................. | 18 | agt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................CCTCAGAGCTGAGAGAGATGTTTCAa...................................................................................................................................................................................................... | 26 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ........................................................................................................................GAGGTTACACCATGAGGCGGCATCAGA....................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................CTAGACCTACCCCTTCAATTTGAAGAttt......................................................................................................................................................................... | 29 | ttt | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................CTAGACCTACCCCTTCAATTTGAAGA......................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |