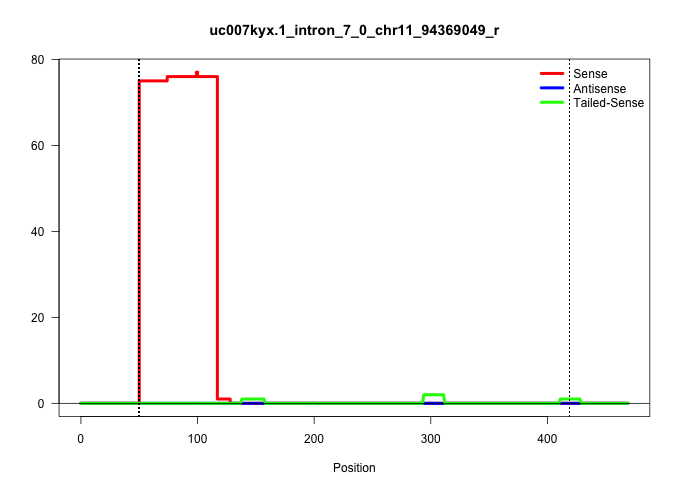
| Gene: Mycbpap | ID: uc007kyx.1_intron_7_0_chr11_94369049_r | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(1) PIWI.ip |
(9) TESTES |
| AGACCTAAAGCAAAACAAGACGCAGAGGTTTTACTTTAACAACCGGGAAGGTATCCTGAAGTTCTCCTTCTTGCTGGCTCAGTGACTGGTGGATCCCCATGTTTAGTTAGTATCTGGCCCATCCCCTCCTCCTCTCATTATGTGTATGGTCCACTATGTCTGTTGAACTCTTGAGAGTGCAGGGGAGGGAGCAGCCTCAGGGAGCACAGACCTGGGGTGGGGATGCCCAGCCATCCCTTGATTGACCCGAGCTTCCCTAGCTCTACATTCAAAAGTCACACAGACCTGGAAACCTTATGCTCCTGAGAAACCAGGACACTGGTCGCCAGATGTGGGCCACTGTGGTGCACTTTCTATAACCAGGTTGGCATCTCAGCCAGCTCCTGCTCCACAGCAGCCTCTGCCTCTGTCTTTCCCAGGTGTGATTCTGCCCGGGGAAACCAAACACTTTACCTTCTTCTTCAAGTCT ...............................................................................................................................................................................................................................................................................................................................(((....((((.((((((..((((((.(((.............(((((((......)))))))..))).))))))..))))..)).))))......)))................................................... ...............................................................................................................................................................................................................................................................................................................................320................................................................................................419................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTATCCTGAAGTTCTCCTTCTTGCTGGCTCAGTGACTGGTGGATCCCCATGT............................................................................................................................................................................................................................................................................................................................................................................... | 52 | 1 | 76.00 | 76.00 | 76.00 | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................................................................................TTATGCTCCTGAGAccgt............................................................................................................................................................. | 18 | ccgt | 2.00 | 0.00 | - | - | - | - | - | - | 1.00 | 1.00 | - |
| ..........................................................................................................................................TATGTGTATGGTCCACatct....................................................................................................................................................................................................................................................................................................................... | 20 | atct | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - |
| ...........................................................................................................................................................................................................................................................................................................................................................................................................................TTTCCCAGGTGTGAcact........................................ | 18 | cact | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 |
| ..........................................................................TGGCTCAGTGACTGGTGGATCCCCAT................................................................................................................................................................................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - |
| ...................................................................................................TGTTTAGTTAGTATCTGGCCCATCCCCTC..................................................................................................................................................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - |
| AGACCTAAAGCAAAACAAGACGCAGAGGTTTTACTTTAACAACCGGGAAGGTATCCTGAAGTTCTCCTTCTTGCTGGCTCAGTGACTGGTGGATCCCCATGTTTAGTTAGTATCTGGCCCATCCCCTCCTCCTCTCATTATGTGTATGGTCCACTATGTCTGTTGAACTCTTGAGAGTGCAGGGGAGGGAGCAGCCTCAGGGAGCACAGACCTGGGGTGGGGATGCCCAGCCATCCCTTGATTGACCCGAGCTTCCCTAGCTCTACATTCAAAAGTCACACAGACCTGGAAACCTTATGCTCCTGAGAAACCAGGACACTGGTCGCCAGATGTGGGCCACTGTGGTGCACTTTCTATAACCAGGTTGGCATCTCAGCCAGCTCCTGCTCCACAGCAGCCTCTGCCTCTGTCTTTCCCAGGTGTGATTCTGCCCGGGGAAACCAAACACTTTACCTTCTTCTTCAAGTCT ...............................................................................................................................................................................................................................................................................................................................(((....((((.((((((..((((((.(((.............(((((((......)))))))..))).))))))..))))..)).))))......)))................................................... ...............................................................................................................................................................................................................................................................................................................................320................................................................................................419................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................................................................CAGCCATCCCTTGATaaca................................................................................................................................................................................................................................... | 19 | aaca | 2.00 | 0.00 | - | 2.00 | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................................................................................................................................................................ATCTCAGCCAGCTCCtta..................................................................................... | 18 | tta | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - |