
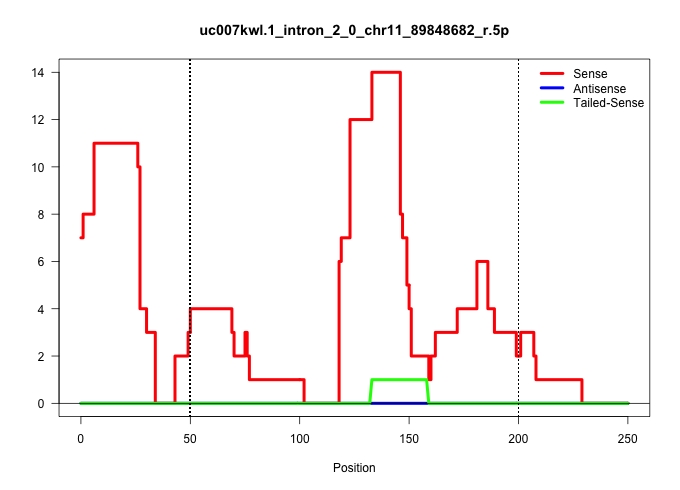
| Gene: Pctp | ID: uc007kwl.1_intron_2_0_chr11_89848682_r.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(11) TESTES |
| TGGTGGCATACTGGGAAGTGAAGTACCCTTTCCCGCTGTCCAACAGAGATGTATCCTTTCTGTACGGCACTCTGTTCTTTGTCTCTCTTCGTGTGCTGCGTTCCCCAGCCGCCGCCCTTACACTGGGATCCACTAACGTCACGGCACTGTCTTGGGTCCTTACAGAACTGTATGCTAATCCTGGAGCTTCTTGTGTGACTTTTGGAGAGTCCAGAGTGTTCTTTCTGGGTGTGACCACTTAGGGCAGGAA ................................................................((((((......................)))))).......(((..(((((((.......)))....((....))..)))).)))..................................................................................................... ..................................................51.................................................................................................150.................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................TACACTGGGATCCACTAACGTCACGGCA........................................................................................................ | 28 | 1 | 6.00 | 6.00 | 1.00 | 2.00 | - | - | - | 2.00 | 1.00 | - | - | - | - |
| ......CATACTGGGAAGTGAAGTACCCTTTCCC........................................................................................................................................................................................................................ | 28 | 1 | 3.00 | 3.00 | - | - | 3.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TGGGATCCACTAACGTCACGGCACTG..................................................................................................... | 26 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TGGGATCCACTAACGTCACGGCACTGTC................................................................................................... | 28 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TAACGTCACGGCACTGTCTTGGGTCC........................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - |
| ...........................................CAGAGATGTATCCTTTCTGTACGGCAC.................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................TGTATCCTTTCTGTACGGCACTCTGTT.............................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...........................................CAGAGATGTATCCTTTCTGTACGGCA..................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...........................................................................TCTTTGTCTCTCTTCGTGTGCTGCGTT.................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................GTATCCTTTCTGTACGGCACTCTGTTC............................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...........................................................................................................................TGGGATCCACTAACGTCACGGCACTGT.................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .GGTGGCATACTGGGAAGTGAAGTACCCTT............................................................................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................TACAGAACTGTATGCTAATCCTGGAG................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .....................................................................................................................................TAACGTCACGGCACTGTCTTGGGTtc........................................................................................... | 26 | tc | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................CAGAACTGTATGCTAATCCTGGAGCTT............................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................................................................................................................ACACTGGGATCCACTAACGTCACGGCAC....................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TTGGAGAGTCCAGAGTGTTCTTTCTGGG..................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................................................................................................TTACAGAACTGTATGCTAATCCTGGAG................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .....................................................................................................................................................................................TGGAGCTTCTTGTGTGACTTTTGGAG........................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................................................................................................................................................................TGCTAATCCTGGAGCTTCTTGTGTGAC................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....................................................................................................................................................................................TGGAGCTTCTTGTGTGACTTTTGGAGA.......................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| TGGTGGCATACTGGGAAGTGAAGTACCCTTTCCCGCTGTCCAACAGAGATGTATCCTTTCTGTACGGCACTCTGTTCTTTGTCTCTCTTCGTGTGCTGCGTTCCCCAGCCGCCGCCCTTACACTGGGATCCACTAACGTCACGGCACTGTCTTGGGTCCTTACAGAACTGTATGCTAATCCTGGAGCTTCTTGTGTGACTTTTGGAGAGTCCAGAGTGTTCTTTCTGGGTGTGACCACTTAGGGCAGGAA ................................................................((((((......................)))))).......(((..(((((((.......)))....((....))..)))).)))..................................................................................................... ..................................................51.................................................................................................150.................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................CTTTCCCGCTGTCCAag................................................................................................................................................................................................................ | 17 | ag | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................................................................................................TATGCTAATCCTGgccc................................................................... | 17 | gccc | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |