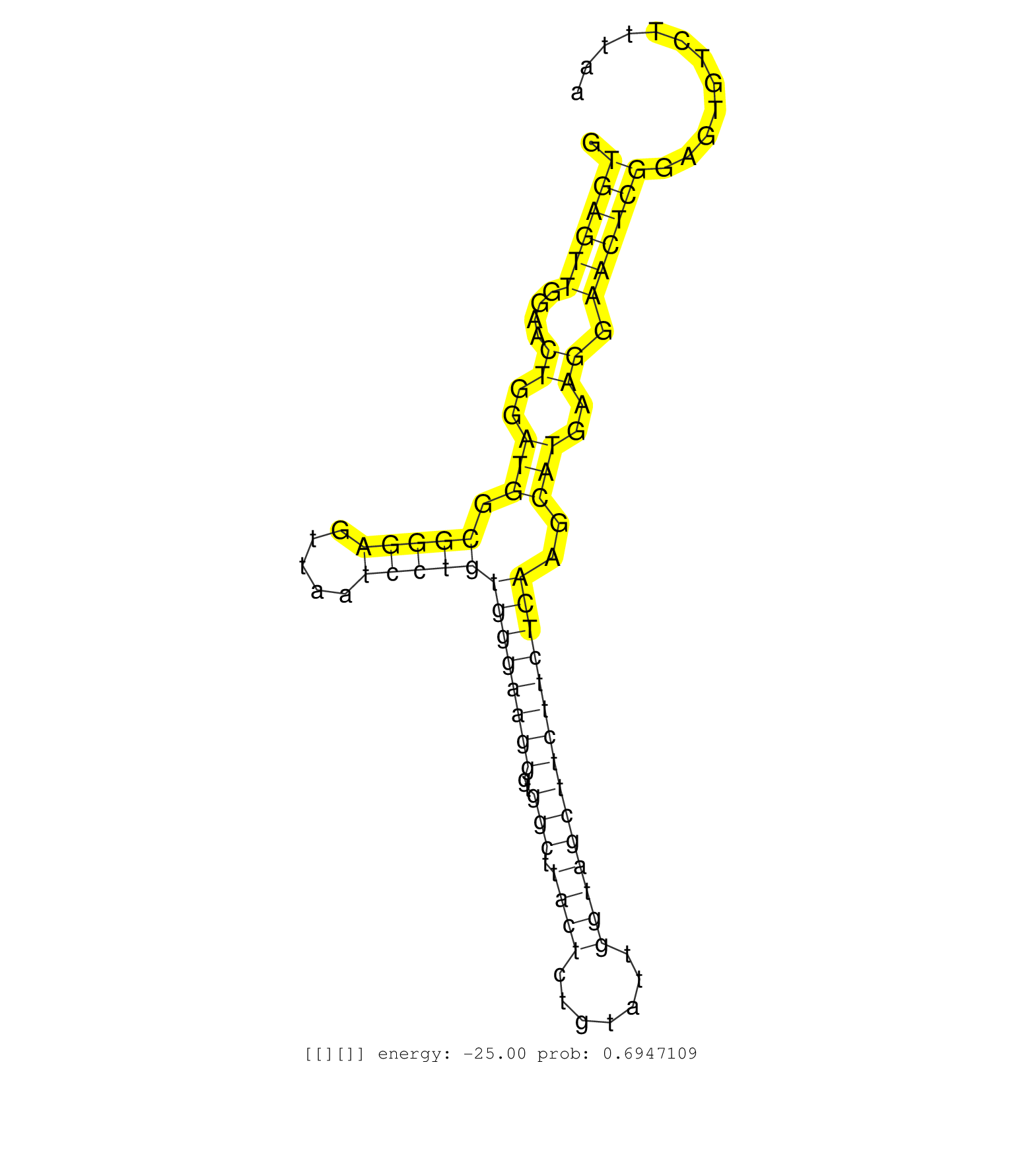

| Gene: Tex14 | ID: uc007ktr.1_intron_2_0_chr11_87308585_f.5p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(1) OVARY |
(3) PIWI.ip |
(3) PIWI.mut |
(15) TESTES |
| GATGAGCCCACCTTCTCTTTCTTCAGTGGCCCCTACATGGTCATGACTAAGTGAGTTGGAACTGGATGGCGGGAGTTAATCCTGTGGGAAGGGTGGCTTACTCTGTATTGGTAGCTTCTTCTCAAGCATGAAGGAACTCGGAGTGTCTTTAAACTAGCCTTTCTTACTGTGGACATTTTTGGCCAGATGATTTCTTGGTTTTTTGTTTGTTTGTTTGTTTTTGTTTTTGTTTTTTGAGACAGGATTTCTC ...................................................((((((....((..(((.(((((.....)))))((((((((..(((.((((.......)))))))))))))))..)))..)).)))))).............................................................................................................. ..................................................51...................................................................................................152................................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGAGTTGGAACTGGATGGCGGGAG............................................................................................................................................................................... | 25 | 1 | 17.00 | 17.00 | 3.00 | 1.00 | 1.00 | 3.00 | 4.00 | 1.00 | - | 1.00 | 1.00 | - | 1.00 | - | - | 1.00 | - | - |
| ..................................................GTGAGTTGGAACTGGATGGCGGGAGT.............................................................................................................................................................................. | 26 | 1 | 8.00 | 8.00 | 2.00 | - | - | 2.00 | - | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTTGGAACTGGATGGCGGGA................................................................................................................................................................................ | 24 | 1 | 7.00 | 7.00 | 2.00 | - | 3.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| .........................................................................................................................TCAAGCATGAAGGAACTCGGAGTGTCT...................................................................................................... | 27 | 1 | 3.00 | 3.00 | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTTGGAACTGGATGGCGGG................................................................................................................................................................................. | 23 | 1 | 3.00 | 3.00 | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................TACTGTGGACATTTTTGGCCAGATGAT........................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...................................................TGAGTTGGAACTGGATGGCGGGAGT.............................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..TGAGCCCACCTTCTCTTTCTTCAGTGG............................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .........................................................................................................................TCAAGCATGAAGGAACTCGGAGT.......................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTTGGAACTGGATGGCGGGAat.............................................................................................................................................................................. | 26 | at | 1.00 | 7.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................TACTGTGGACATTTTTGGCCAGA............................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................TCTTCTCAAGCATGAAGGAACTCGGA............................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................TTCTCAAGCATGAAGGAACTCGGAGTGT........................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTTGGAACTGGATGGCGGGAGTT............................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................TTCTTACTGTGGACATTTTTGGCCA................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..TGAGCCCACCTTCTCTTTCTTCAGTGGC............................................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| GATGAGCCCACCTTCTCTTTCTTCAGTGGCCCCTACATGGTCATGACTAAGTGAGTTGGAACTGGATGGCGGGAGTTAATCCTGTGGGAAGGGTGGCTTACTCTGTATTGGTAGCTTCTTCTCAAGCATGAAGGAACTCGGAGTGTCTTTAAACTAGCCTTTCTTACTGTGGACATTTTTGGCCAGATGATTTCTTGGTTTTTTGTTTGTTTGTTTGTTTTTGTTTTTGTTTTTTGAGACAGGATTTCTC ...................................................((((((....((..(((.(((((.....)))))((((((((..(((.((((.......)))))))))))))))..)))..)).)))))).............................................................................................................. ..................................................51...................................................................................................152................................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|