
| Gene: ni-2 | ID: uc007ksx.1_intron_3_0_chr11_86478292_r.3p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(3) PIWI.ip |
(11) TESTES |
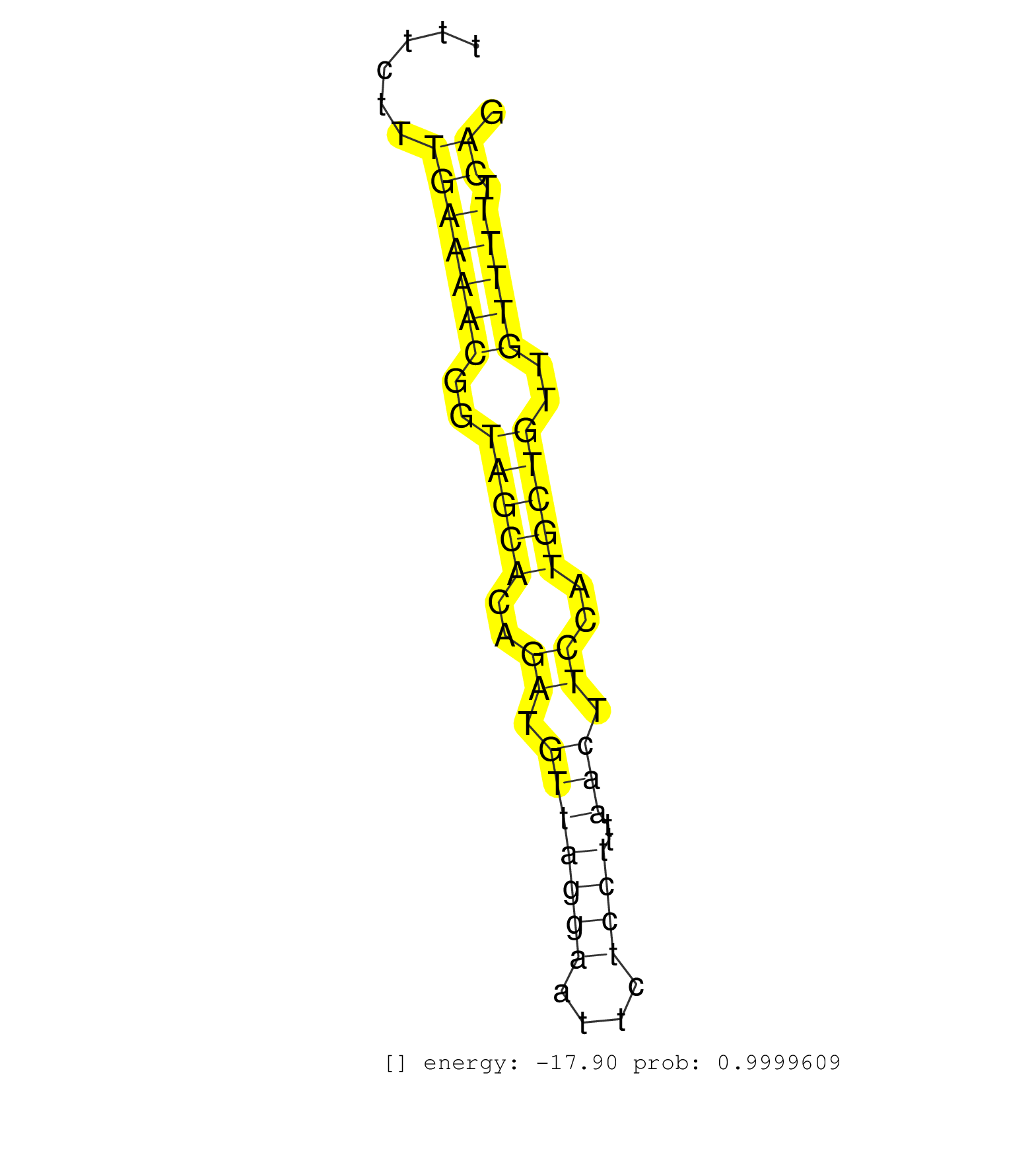
| ACAGAAGGCTGATCTGAGTTTGGTACTTTGAACTCCTTAAATTTGCATGGTCTTAATGTTAAATCTCATTTATACTTTTGAGGTATTTTTATGCTATTAGAGATACAAGACAGCAAGGTGGGTCGATTTTTTTTTTTCTTTGAAAACGGTAGCACAGATGTTAGGAATTCTCCTTTAACTTCCATGCTGTTGTTTTTCAGACCCCTCTTCAGTTCATGAGAAGAAGAGAAGGGATCGGGAAGAAAGACAG ............................................................................................................................................(((((((..(((((..((.(((((((....))))..))).))..)))))..))))).))................................................... ......................................................................................................................................135..............................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................TTGAAAACGGTAGCACAGATGT......................................................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - |
| ...........................................................................................................................................TTGAAAACGGTAGCACAGATGTTAGGAAT.................................................................................. | 29 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TGAAAACGGTAGCACAGATGTT........................................................................................ | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 |
| .............................................................................................................................................GAAAACGGTAGCACAGATGTT........................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGAGAAGAAGAGAAGGGATCGGGAAG........ | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ............................................................................................................................................TGAAAACGGTAGCACAGATGT......................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...................................................................................................................................................................................TTCCATGCTGTTGTTTTTCAGt................................................. | 22 | t | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...............................................................................................................................................................................................................TTCAGTTCATGAGAAGAAGAGAAGGG................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TTGAAAACGGTAGCACAGATGTa........................................................................................ | 23 | a | 1.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGAGAAGAAGAGAAGGGATCGGGAAGt....... | 27 | t | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ACAGAAGGCTGATCTGAGTTTGGTACTTTGAACTCCTTAAATTTGCATGGTCTTAATGTTAAATCTCATTTATACTTTTGAGGTATTTTTATGCTATTAGAGATACAAGACAGCAAGGTGGGTCGATTTTTTTTTTTCTTTGAAAACGGTAGCACAGATGTTAGGAATTCTCCTTTAACTTCCATGCTGTTGTTTTTCAGACCCCTCTTCAGTTCATGAGAAGAAGAGAAGGGATCGGGAAGAAAGACAG ............................................................................................................................................(((((((..(((((..((.(((((((....))))..))).))..)))))..))))).))................................................... ......................................................................................................................................135..............................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|