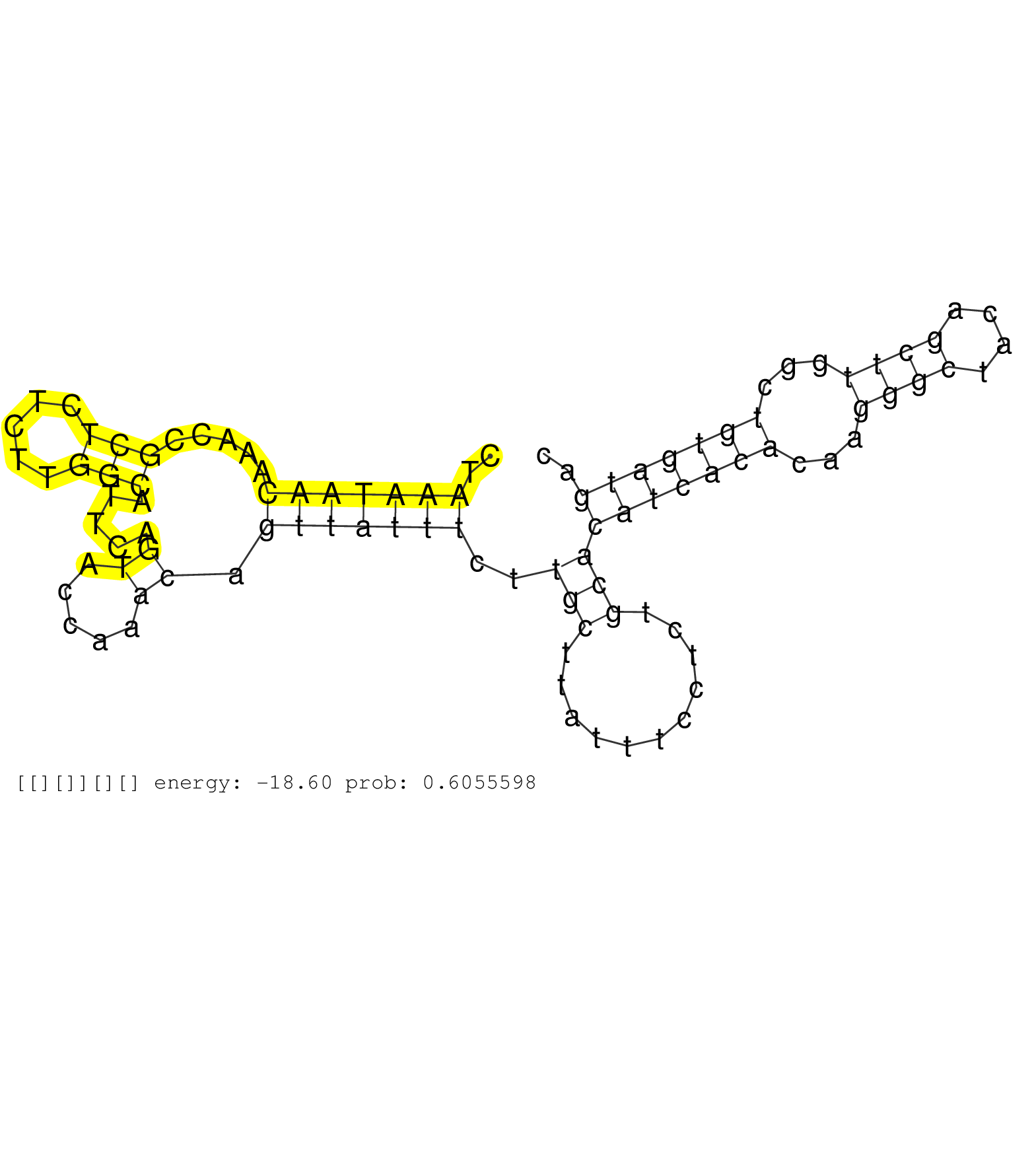
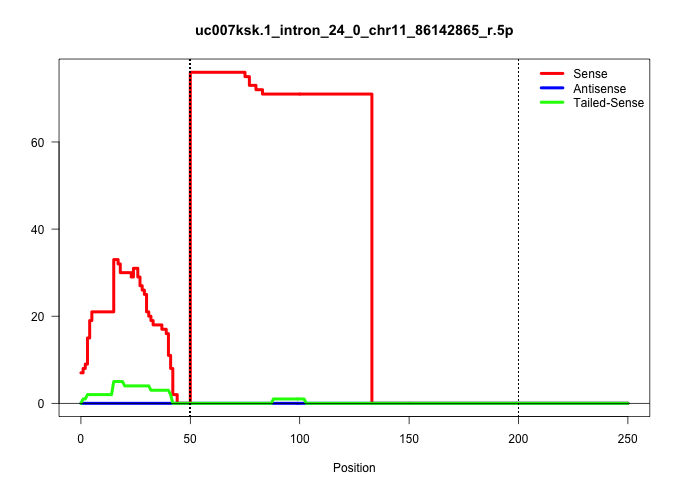
| Gene: Med13 | ID: uc007ksk.1_intron_24_0_chr11_86142865_r.5p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(24) TESTES |
| CGATATGGATTGGGAAGACGATTCTTTGGCTGCGGTGGAAGTTCTTGTTGGTATGGATTCTGATACTAGTCTGAAGGGGGTCCTGTGTGTTTAGATTTTACTAAATAACAAACCGCTCTCTTGGCATTCAGTACCAAACAGTTATTTCTTGCTTATTTCCTCTGCACATCACACAAGGGCTACAGCTTGGCTGTGATGACTGTCTGCATGTTCTTCCTGTACAGTCCAGGTTAAGAAGCACCTAGGGTCT ......................................................................................................(((((((.....(((.....))).....((.....)).)))))))..(((...........)))(((((((...((((....))))...))))))).................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTATGGATTCTGATACTAGTCTGAAGGGGGTCCTGTGTGTTTAGATTTTACT.................................................................................................................................................... | 52 | 1 | 74.00 | 74.00 | 74.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............AAGACGATTCTTTGGCTGCGG....................................................................................................................................................................................................................... | 21 | 1 | 10.00 | 10.00 | - | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTATGGATTCTGATACTAGTCTGAAGGGGGTC........................................................................................................................................................................ | 32 | 1 | 8.00 | 8.00 | - | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................GTATGGATTCTGATACTAGTCTGAAGGGGG.......................................................................................................................................................................... | 30 | 1 | 6.00 | 6.00 | - | - | - | - | 1.00 | - | - | 4.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............AGACGATTCTTTGGCTGCGGTGGAA.................................................................................................................................................................................................................. | 25 | 1 | 5.00 | 5.00 | - | - | 1.00 | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...............AGACGATTCTTTGGCTGCGGTGGAAGT................................................................................................................................................................................................................ | 27 | 1 | 4.00 | 4.00 | - | - | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...TATGGATTGGGAAGACGATTCTTTGGC............................................................................................................................................................................................................................ | 27 | 1 | 3.00 | 3.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..ATATGGATTGGGAAGACGATT................................................................................................................................................................................................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............AGACGATTCTTTGGCTGCGGTGGAAG................................................................................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTATGGATTCTGATACTAGTCTGAAGG............................................................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TGGATTGGGAAGACGATTCTTTGGCTGC......................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...TATGGATTGGGAAGACGATTCTTTGGCaa.......................................................................................................................................................................................................................... | 29 | aa | 1.00 | 3.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .GATATGGATTGGGAAGACa...................................................................................................................................................................................................................................... | 19 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....ATGGATTGGGAAGACGATT................................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TGGATTGGGAAGACGATTCTTTGGC............................................................................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................CTTTGGCTGCGGTGGAAGTTC.............................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............AGACGATTCTTTGGCTGCGGTGGAAtg................................................................................................................................................................................................................ | 27 | tg | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................TTGGCTGCGGTGGAAGT................................................................................................................................................................................................................ | 17 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTATGGATTCTGATACTAGTCTGAA............................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................GTTTAGATTTTACga................................................................................................................................................... | 15 | ga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TATGGATTGGGAAGACGATT................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....ATGGATTGGGAAGACGATTCTTTGGCTG.......................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................GGTATGGATTCTGATACTAGTCTGAAG.............................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....ATGGATTGGGAAGACGATTCTTTGGCTGCGGTG..................................................................................................................................................................................................................... | 33 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTATGGATTCTGATACTAGTCTGAAGGGGGTCC....................................................................................................................................................................... | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............AGACGATTCTTTGGCTGCGGTGGA................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................TTTGGCTGCGGTGGAAGTTC.............................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................TTTGGCTGCGGTGGAAGT................................................................................................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................CTTTGGCTGCGGTGGAAG................................................................................................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....ATGGATTGGGAAGACGATTCTTTGGCT........................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TATGGATTGGGAAGACGATTCTTTGG............................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................GTATGGATTCTGATACTAGTCTGAAGGG............................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .GATATGGATTGGGAAGA........................................................................................................................................................................................................................................ | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............AGACGATTCTTTGGCTGCGGTGGAAGa................................................................................................................................................................................................................ | 27 | a | 1.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TATGGATTGGGAAGACGATTCTT................................................................................................................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...............AGACGATTCTTTGGCTGCGGTGGAAt................................................................................................................................................................................................................. | 26 | t | 1.00 | 5.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .GATATGGATTGGGAAGACGATTCTTTGGC............................................................................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| CGATATGGATTGGGAAGACGATTCTTTGGCTGCGGTGGAAGTTCTTGTTGGTATGGATTCTGATACTAGTCTGAAGGGGGTCCTGTGTGTTTAGATTTTACTAAATAACAAACCGCTCTCTTGGCATTCAGTACCAAACAGTTATTTCTTGCTTATTTCCTCTGCACATCACACAAGGGCTACAGCTTGGCTGTGATGACTGTCTGCATGTTCTTCCTGTACAGTCCAGGTTAAGAAGCACCTAGGGTCT ......................................................................................................(((((((.....(((.....))).....((.....)).)))))))..(((...........)))(((((((...((((....))))...))))))).................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|