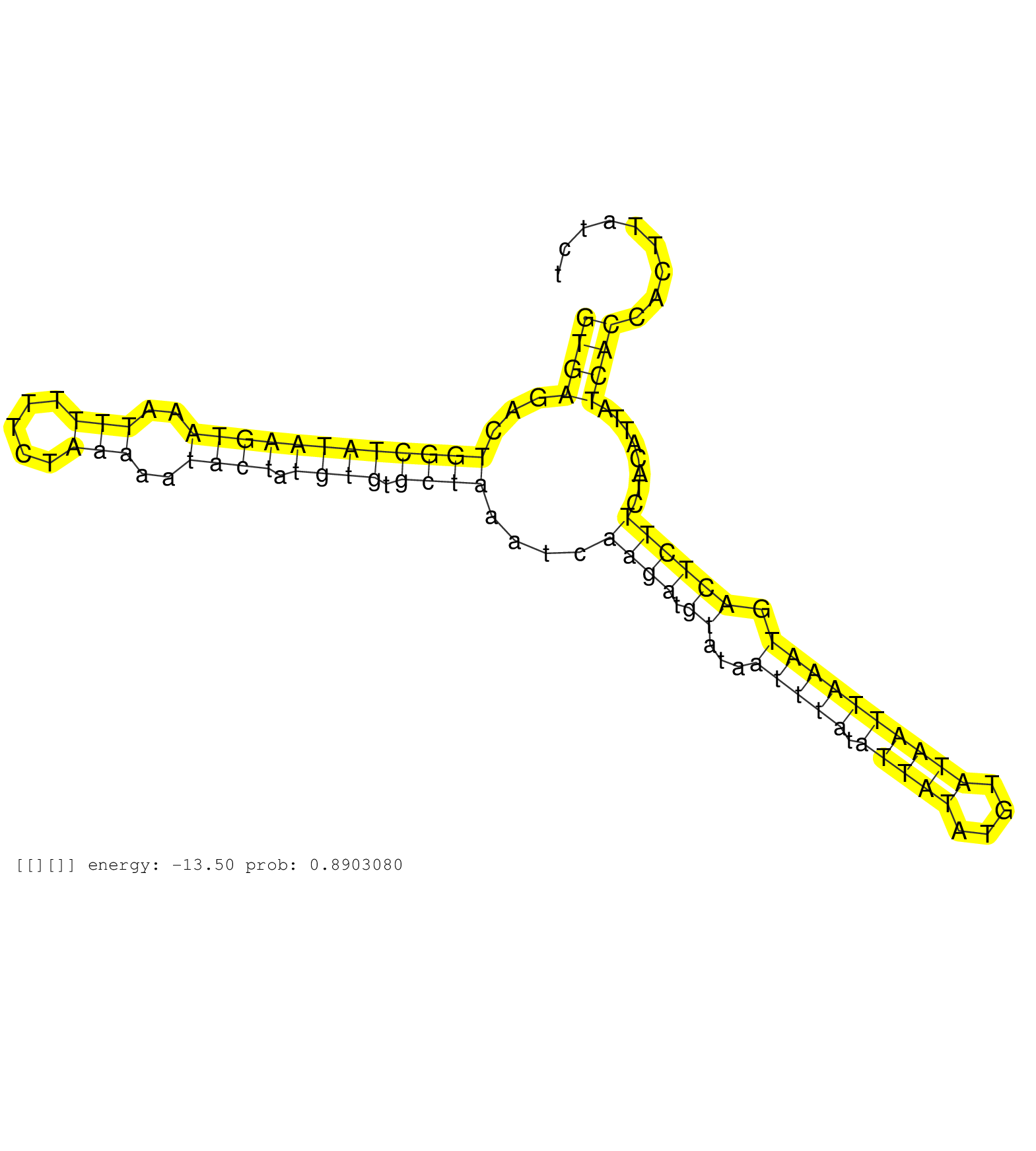

| Gene: Usp32 | ID: uc007krh.1_intron_24_0_chr11_84864777_r.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(4) OTHER.mut |
(5) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(24) TESTES |
| ATTTGTCAGACATTGTAGAAAGGATATTGAATGCACATGATACCACAAAGGTGAGACTGGCTATAAGTAAATTTTTTCTAAAAATACTATGTGTGCTAAATCAAGATGTATAATTTATATTATATGTATAATTAAATGACTCTTCTACATTATCACCACTTATCTGGTCCAGTGGTAAAATACCTTTACCTTTACATCGTCTTTAAAAATAGAGGTCTATTCCATCCTCAACAGCAACTCAGTGAAGTCT ..................................................((((...((((((((((((..(((.....)))..)))).)))).))))....((((.((...(((((.(((((....)))))))))).))))))........)))).............................................................................................. ..................................................51................................................................................................................165................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................TTATATGTATAATTAAATGACTCTTCTACATTATCACCACTTATCTGGTCCA............................................................................... | 52 | 1 | 46.00 | 46.00 | 46.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TGTCAGACATTGTAGAAAGGATATTGAA........................................................................................................................................................................................................................... | 28 | 1 | 8.00 | 8.00 | - | - | - | - | 2.00 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...............TAGAAAGGATATTGAATGCACATGATACC.............................................................................................................................................................................................................. | 29 | 1 | 4.00 | 4.00 | - | 2.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TGTCAGACATTGTAGAAAGGATATTGA............................................................................................................................................................................................................................ | 27 | 1 | 4.00 | 4.00 | - | - | - | - | 2.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........ACATTGTAGAAAGGATATTGAATGCACAT.................................................................................................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........GACATTGTAGAAAGGATATTGAATGCACAT.................................................................................................................................................................................................................... | 30 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........ATTGTAGAAAGGATATTGAATGCACA..................................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................GAAAGGATATTGAATGCACATGATACC.............................................................................................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........GACATTGTAGAAAGGATATTGAAT.......................................................................................................................................................................................................................... | 24 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........ATTGTAGAAAGGATATTGAATGCACAa.................................................................................................................................................................................................................... | 27 | a | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............TTGTAGAAAGGATATTGAATGCACATG................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...........ATTGTAGAAAGGATATTGAATGCACATGA.................................................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......AGACATTGTAGAAAGGATATTGAATGC........................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........ATTGTAGAAAGGATATTGAATGC........................................................................................................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ................AGAAAGGATATTGAATGCACATGATACC.............................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................GTGAGACTGGCTATAAGTAAATTTTTTCTA.......................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................ATCTGGTCCAGTGGTAg........................................................................ | 17 | g | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TCAGACATTGTAGAAAGGATATTGAATGC........................................................................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TCAGACATTGTAGAAAGGA.................................................................................................................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............TAGAAAGGATATTGAATGCACATGAT................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............TTGTAGAAAGGATATTGAATGCACAT.................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................GAAAGGATATTGAATGCACATGA.................................................................................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............TTGTAGAAAGGATATTGAATGCAC...................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............TAGAAAGGATATTGAATGC........................................................................................................................................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........GACATTGTAGAAAGGATATTGAATGC........................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................AAAGGTGAGACTGGCTATAAGTAAATTT................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .......AGACATTGTAGAAAGGATATT.............................................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...TGTCAGACATTGTAGAAAGGATATTGtt........................................................................................................................................................................................................................... | 28 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...........ATTGTAGAAAGGATATTGAATGCAC...................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....GTCAGACATTGTAGAAAGGATATTGAA........................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................AGGATATTGAATGCACATGATA................................................................................................................................................................................................................ | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............TGTAGAAAGGATATTGAATGCACATGttt................................................................................................................................................................................................................ | 29 | ttt | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............TTGTAGAAAGGATATTGAATGCgcat.................................................................................................................................................................................................................... | 26 | gcat | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ATTTGTCAGACATTGTAGAAAGGATATTGAATGCACATGATACCACAAAGGTGAGACTGGCTATAAGTAAATTTTTTCTAAAAATACTATGTGTGCTAAATCAAGATGTATAATTTATATTATATGTATAATTAAATGACTCTTCTACATTATCACCACTTATCTGGTCCAGTGGTAAAATACCTTTACCTTTACATCGTCTTTAAAAATAGAGGTCTATTCCATCCTCAACAGCAACTCAGTGAAGTCT ..................................................((((...((((((((((((..(((.....)))..)))).)))).))))....((((.((...(((((.(((((....)))))))))).))))))........)))).............................................................................................. ..................................................51................................................................................................................165................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|