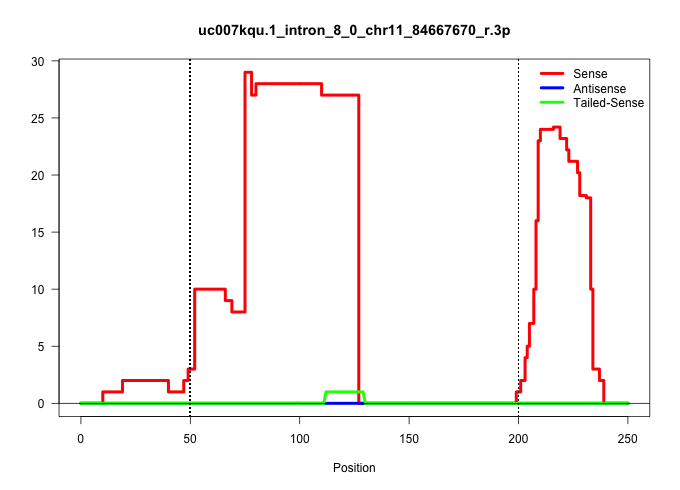
| Gene: Ggnbp2 | ID: uc007kqu.1_intron_8_0_chr11_84667670_r.3p | SPECIES: mm9 |
 |
|
(3) OTHER.mut |
(2) PIWI.ip |
(1) TDRD1.ip |
(18) TESTES |
| AGCAGTGTCAGTTCAGAGAAGTTAAGTACAGATAAGAGAAGTAGTGAAGACCACAGGAAGGACAGCAAGTGTAGGATCATTTTCCATTATGGACCTTTCCAGGGAACAGCCAGGTAAAACCAAGCAATCCTTTATTCTGCCTTTTTAAACCTTTCGCCTCATGTATGCCTCCTTTTCACTAATATTTATTCTTTTTACAGAGGATGTTGGATGGATGTTTGGGAACTCATGTCCCAGGAATGCAGGGATG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | mjTestesWT2() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................ATCATTTTCCATTATGGACCTTTCCAGGGAACAGCCAGGTAAAACCAAGCAA........................................................................................................................... | 52 | 1 | 27.00 | 27.00 | 27.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................GATGGATGTTTGGGAACTCATGTCC................ | 25 | 1 | 7.00 | 7.00 | - | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................ACAGGAAGGACAGCAAGTGTAGG............................................................................................................................................................................... | 23 | 1 | 6.00 | 6.00 | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TGGATGGATGTTTGGGAACTCATGTC................. | 26 | 1 | 3.00 | 3.00 | - | - | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................GGATGGATGTTTGGGAACTCATGTC................. | 25 | 1 | 3.00 | 3.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................GTTGGATGGATGTTTGGGAACTCATGTC................. | 28 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................GGTAAAACCAAGCAgtca........................................................................................................................ | 18 | gtca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ................................................................................................................................................................................................................GGATGGATGTTTGGGAACTCATGTCCCAGGA........... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ....................................................ACAGGAAGGACAGCAAGTGTAGGATC............................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...................AGTTAAGTACAGATAAGAGAAGTAGT............................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................ATGTTGGATGGATGTTTGGG........................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .......................................................................................................................................................................................................GAGGATGTTGGATGGATGTTTGGGAACTC...................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TGTTGGATGGATGTTTGG............................ | 18 | 2 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........GTTCAGAGAAGTTAAGTACAGATAAGAGAA.................................................................................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................TTTCCATTATGGACCTTTCCAGGGAACAGC............................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................................GAAGACCACAGGAAGGACAGC........................................................................................................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ................................................................................................................................................................................................................GGATGGATGTTTGGGAACTCATGTCCCAG............. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................ATGGATGTTTGGGAACTCATGTCCCAGGA........... | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................ATGTTGGATGGATGTTTGGGAACTC...................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .........................................................................................................................................................................................................GGATGTTGGATGGATGTT............................... | 18 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................GGATGGATGTTTGGGAACT....................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................ACCACAGGAAGGACAGCAAGTGTAGGATC............................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...............................................AGACCACAGGAAGGACAGCAAG..................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................GTTTGGGAACTCATG................... | 15 | 5 | 0.20 | 0.20 | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGCAGTGTCAGTTCAGAGAAGTTAAGTACAGATAAGAGAAGTAGTGAAGACCACAGGAAGGACAGCAAGTGTAGGATCATTTTCCATTATGGACCTTTCCAGGGAACAGCCAGGTAAAACCAAGCAATCCTTTATTCTGCCTTTTTAAACCTTTCGCCTCATGTATGCCTCCTTTTCACTAATATTTATTCTTTTTACAGAGGATGTTGGATGGATGTTTGGGAACTCATGTCCCAGGAATGCAGGGATG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | mjTestesWT2() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................ACCTTTCCAGGGAACgcgc............................................................................................................................................... | 19 | gcgc | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |