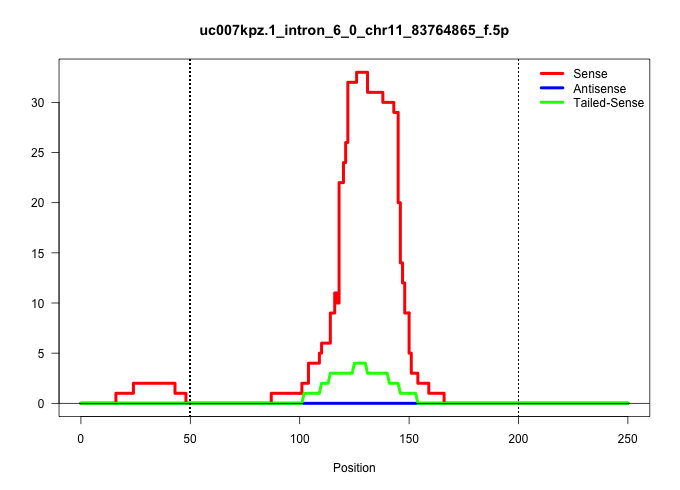
| Gene: Ddx52 | ID: uc007kpz.1_intron_6_0_chr11_83764865_f.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(2) OTHER.mut |
(3) PIWI.ip |
(1) PIWI.mut |
(20) TESTES |
| CGACTGATCTACTTGTTGAAGCAGGATCCACCAGGGATTGACCTAACAAAGTACTATGTTCTCATGTCTTTCATTGACTCACTTATTTTATTAGTTTTCTTGTTACTGTGACTGATTATTTGACAAAGAGAAATTGAAGGAACGGAGGAAGGCCTGTGTGTGATTCATTGTGGCATTGGGGCAGGAAGGTGAGGCCTGGACTCCTTGCATCCACAGTCAGAAGCAGGACAGCGATGGCTGTTGACCCTTA .........................................................................................................((((..((((((.(((......))).))))..))..))))......((((..((((.((.....)).))))..))))....(((.....)))..................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................TTTGACAAAGAGAAATTGAAGGAACGG......................................................................................................... | 27 | 1 | 6.00 | 6.00 | - | 2.00 | - | - | - | - | - | 1.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................TTTGACAAAGAGAAATTGAAGGAACGGA........................................................................................................ | 28 | 1 | 4.00 | 4.00 | 1.00 | - | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................ACAAAGAGAAATTGAAGGAACGGAGGAA.................................................................................................... | 28 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - |
| ..........................................................................................................................ACAAAGAGAAATTGAAGGAACGGAGG...................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ......................................................................................................................TTTGACAAAGAGAAATTGAAGGAACGGAG....................................................................................................... | 29 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................ATTATTTGACAAAGAGAAATTGAAGGAACGG......................................................................................................... | 31 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................ACTGTGACTGATTATTTGACAAAGAGA....................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................ACTGATTATTTGACAAAGAGAAATTGAAGG.............................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ........................................................................................................................TGACAAAGAGAAATTGAAGGAACGGAGG...................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................ATTATTTGACAAAGAGAAATTGAAGGAAC........................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................ATTGAAGGAACGGAGGAAGGCCTGTGT........................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................GATCCACCAGGGATTGACCTAACA.......................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .........................................................................................................................GACAAAGAGAAATTGAAGGAACGGAGGAAG................................................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TTACTGTGACTGATTATTTGACAAAGAGt....................................................................................................................... | 29 | t | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................TTATTAGTTTTCTTGTTACTGTGACTGATT..................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................TATTTGACAAAGAGAAATTGAAGGAACGGA........................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................TGAAGCAGGATCCACCAGGGATTGACC............................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................AAGAGAAATTGAAGGAACGGAGGAAGGCt................................................................................................ | 29 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................GACTGATTATTTGACAAAGAGAAATTGAA................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................GACAAAGAGAAATTGAAGGAACGGAGGAA.................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ........................................................................................................................TGACAAAGAGAAATTGAAGGAACGGA........................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..............................................................................................................................AGAGAAATTGAAGGAACGGAGGAAGGCC................................................................................................ | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................GTTACTGTGACTGATTATTTGACAAAGAGAA...................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................AACGGAGGAAGGCCTGTGTGTGATTC.................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................ACTGATTATTTGACAAAGAGAAATTGAAGGt............................................................................................................. | 31 | t | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................ACAAAGAGAAATTGAAGGAACGGAGGAAG................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................ATTATTTGACAAAGAGAAATTGAAGGAACGGt........................................................................................................ | 32 | t | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TATTTGACAAAGAGAAATTGAAGGAACGG......................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CGACTGATCTACTTGTTGAAGCAGGATCCACCAGGGATTGACCTAACAAAGTACTATGTTCTCATGTCTTTCATTGACTCACTTATTTTATTAGTTTTCTTGTTACTGTGACTGATTATTTGACAAAGAGAAATTGAAGGAACGGAGGAAGGCCTGTGTGTGATTCATTGTGGCATTGGGGCAGGAAGGTGAGGCCTGGACTCCTTGCATCCACAGTCAGAAGCAGGACAGCGATGGCTGTTGACCCTTA .........................................................................................................((((..((((((.(((......))).))))..))..))))......((((..((((.((.....)).))))..))))....(((.....)))..................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|