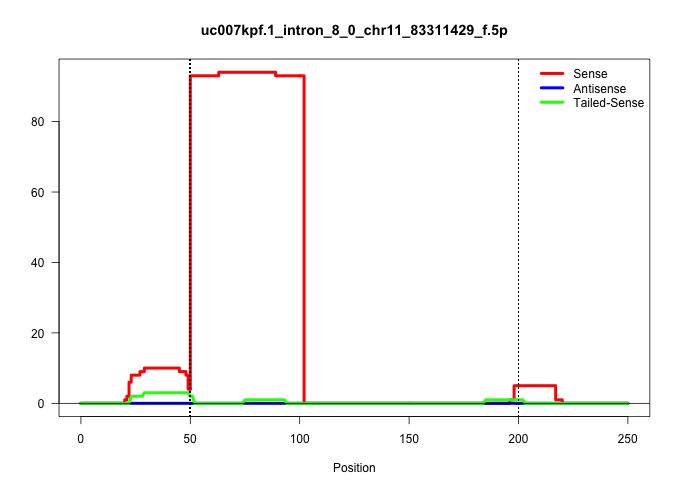
| Gene: Taf15 | ID: uc007kpf.1_intron_8_0_chr11_83311429_f.5p | SPECIES: mm9 |
 |
|
(4) OTHER.mut |
(2) PIWI.ip |
(2) PIWI.mut |
(15) TESTES |
| CTTTTCTTCTCCCATAGGTCACAGGGATTATGGACCCAGACCAGATGCTGGTAAGATTCTTGATAGTTTGGTTTCCTACCTCTGGAAGAGGAATTAATGTGTTTGGAGGGAGGTTTAATTTAATTTAATTTAATTTAATTTAATTTAATGGTGCTGCCAGATCTGGGGAATTTGACTTAAAATAATAATTTAGTGATCTGATTAGAAAGCTTGCTGGAGAAACTGCTGTGTTTGTGCTTTGTTGCTTGAA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGATTCTTGATAGTTTGGTTTCCTACCTCTGGAAGAGGAATTAATGTGT.................................................................................................................................................... | 52 | 1 | 93.00 | 93.00 | 93.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................TGATTAGAAAGCTTGCTGG................................. | 19 | 1 | 4.00 | 4.00 | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................AGGGATTATGGACCCAGACCAGATGCT......................................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - |
| .............................ATGGACCCAGACCAGATGCTGaa...................................................................................................................................................................................................... | 23 | aa | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................AGGGATTATGGACCCAGACCAGA............................................................................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................AGGGATTATGGACCCAGACCAGATGCTG........................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................CAGGGATTATGGACCCAGACCAGATGCT......................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................GGGATTATGGACCCAGACCAGATGCTt........................................................................................................................................................................................................ | 27 | t | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................TTATGGACCCAGACCAGATGC.......................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .......................GGGATTATGGACCCAGACCAGATGCTGat...................................................................................................................................................................................................... | 29 | at | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TAGTTTGGTTTCCTACCTCTGGAAGA................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .......................GGGATTATGGACCCAGACCAGATGCT......................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................CTACCTCTGGAAGAGcagg............................................................................................................................................................ | 19 | cagg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................ATGGACCCAGACCAGATGCTG........................................................................................................................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ....................ACAGGGATTATGGACCCAGACCAGATGCTG........................................................................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................GGGATTATGGACCCAGACCAGATGCTG........................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................TCTGATTAGAAAGCTTGCTGGAGA.............................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................................................................................................................................................................................TAATTTAGTGATCTaatg............................................... | 18 | aatg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| CTTTTCTTCTCCCATAGGTCACAGGGATTATGGACCCAGACCAGATGCTGGTAAGATTCTTGATAGTTTGGTTTCCTACCTCTGGAAGAGGAATTAATGTGTTTGGAGGGAGGTTTAATTTAATTTAATTTAATTTAATTTAATTTAATGGTGCTGCCAGATCTGGGGAATTTGACTTAAAATAATAATTTAGTGATCTGATTAGAAAGCTTGCTGGAGAAACTGCTGTGTTTGTGCTTTGTTGCTTGAA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............TCACAGGGATTATGgttc.......................................................................................................................................................................................................................... | 18 | gttc | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |