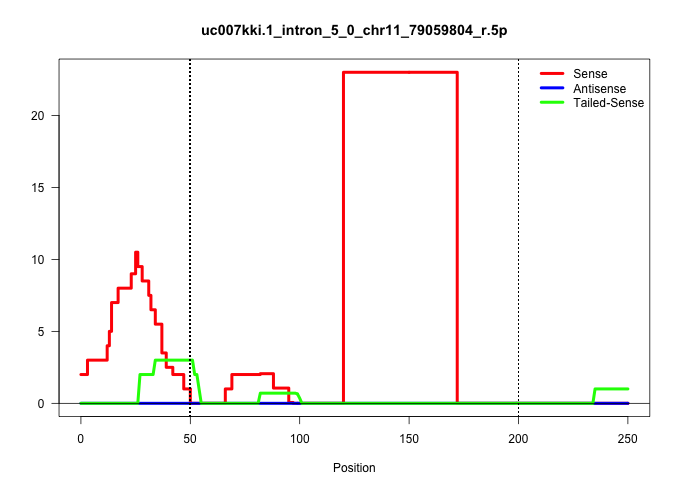
| Gene: Wsb1 | ID: uc007kki.1_intron_5_0_chr11_79059804_r.5p | SPECIES: mm9 |
 |
|
(4) OTHER.mut |
(1) PIWI.mut |
(1) TDRD1.ip |
(15) TESTES |
| TGCCACAGGATTAAACAATGGTCGCATCAAAATCTGGGATGTATATACAGGTATGGATTCATATTGGAAGTGAACCTGGAAAGTGAGAATTAATGTGTCTTTATAATTGTCCTAAGTCTTATAATAATTTATAATTTTAATCTAGGAGAATAATAAGAGTTTATACCACTTGATAAGATAAAATATGCAAAGCAGGTTTGGGTCTTATTTTAGTTAACTATAAACTAATATTTTAGTTTCTGACTCAGCA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................ATAATAATTTATAATTTTAATCTAGGAGAATAATAAGAGTTTATACCACTTG.............................................................................. | 52 | 1 | 23.00 | 23.00 | 23.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............ACAATGGTCGCATCAAAATCTGG..................................................................................................................................................................................................................... | 23 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................CAAAATCTGGGATGTATATACAGGaaaa................................................................................................................................................................................................... | 28 | aaaa | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............AAACAATGGTCGCATCAAA........................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............AACAATGGTCGCATCAAAATCTGGGA................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...CACAGGATTAAACAATGGTCGCATCAAAA.......................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................TGGGATGTATATACAGGaaa.................................................................................................................................................................................................... | 20 | aaa | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................GAAGTGAACCTGGAAAGTGAGA.................................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .......................GCATCAAAATCTGGGATGTATATACAG........................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................ATCAAAATCTGGGATGTATATA........................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................CAAAATCTGGGATGTATATACAGGa...................................................................................................................................................................................................... | 25 | a | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................GTGAACCTGGAAAGTGAGAATTAATG........................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................GTTTCTGACTCAGCctta | 18 | ctta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .................ATGGTCGCATCAAAATC........................................................................................................................................................................................................................ | 17 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................ATCAAAATCTGGGATGT................................................................................................................................................................................................................ | 17 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................GTGAGAATTAATGTGaatt..................................................................................................................................................... | 19 | aatt | 0.35 | 0.06 | - | - | 0.06 | 0.18 | - | - | - | - | - | - | - | - | - | 0.06 | 0.06 |
| ..................................................................................GTGAGAATTAATGTGaat...................................................................................................................................................... | 18 | aat | 0.29 | 0.06 | - | - | - | 0.18 | - | - | - | - | - | - | - | - | - | 0.12 | - |
| ..................................................................................GTGAGAATTAATGTGaa....................................................................................................................................................... | 17 | aa | 0.06 | 0.06 | - | - | 0.06 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................GTGAGAATTAATGTG......................................................................................................................................................... | 15 | 17 | 0.06 | 0.06 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.06 |
| TGCCACAGGATTAAACAATGGTCGCATCAAAATCTGGGATGTATATACAGGTATGGATTCATATTGGAAGTGAACCTGGAAAGTGAGAATTAATGTGTCTTTATAATTGTCCTAAGTCTTATAATAATTTATAATTTTAATCTAGGAGAATAATAAGAGTTTATACCACTTGATAAGATAAAATATGCAAAGCAGGTTTGGGTCTTATTTTAGTTAACTATAAACTAATATTTTAGTTTCTGACTCAGCA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT2() Testes Data. (testes) |
|---|