
| Gene: Nek8 | ID: uc007kih.1_intron_9_0_chr11_77984455_r | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(4) PIWI.ip |
(13) TESTES |
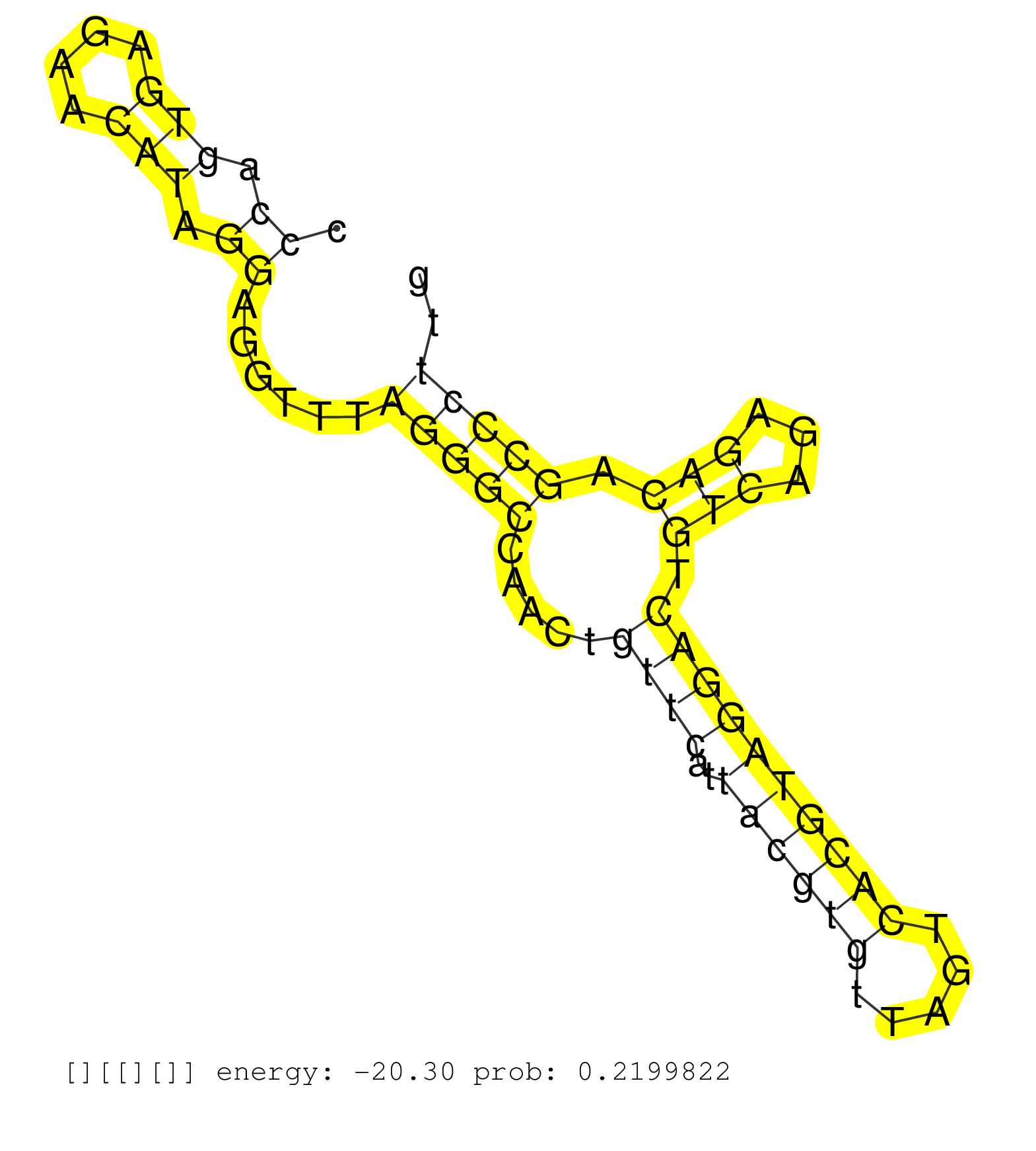
| CGGGCCCTACTCAACATCCACACCGATGTGGGCAGCGTGCGCATGCGGAGGCCTGTGCAGGGAGATGGGTCCTGGGGCGGGCACCCAGTGAGAACATAGGAGGTTTAGGGCCAACTGTTCATTACGTGTTAGTCACGTAGGACTGTCAGAGACAGCCCTTGTCCTGAGGCCCCTTTGTATTCCGCAGGGCAGAGAAGTCCCTGACCCCAGGACCACCTATAGCCTCTGGCAGCACAG ....................................................................................((.(((....))).))......(((((.....((((..((((((.....)))))))))).(((...))).))))).............................................................................. ...................................................................................84...........................................................................161.......................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................TAGTCACGTAGGACTGTCAGAGACAGCC................................................................................ | 28 | 1 | 8.00 | 8.00 | 3.00 | - | 3.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - |
| .................................................................................................................................TAGTCACGTAGGACTGTCAGAGACAGC................................................................................. | 27 | 1 | 7.00 | 7.00 | 2.00 | - | - | 3.00 | - | 1.00 | - | 1.00 | - | - | - | - | - |
| ........................................................................................TGAGAACATAGGAGGTTTAGGGCCAAC.......................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - |
| ......................................................................................................................TCATTACGTGTTAGTCACGTAGGACTGT........................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................TTAGGGCCAACTGTTCATTACGTGTT........................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...........................................TGCGGAGGCCTGTGCAGGGAGATGGGT....................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ........................................................................................TGAGAACATAGGAGGTTTAGGGCCAAt.......................................................................................................................... | 27 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................................................................................................................TTACGTGTTAGTCACGTAGGACTGTCAGAt...................................................................................... | 30 | t | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ................................................................................................................................TTAGTCACGTAGGACTGTCAGAGACAGCC................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................TGCGGAGGCCTGTGCAGGGAGATGGG........................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...................................................................................................................................GTCACGTAGGACTGTCAGAGACAGCC................................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TAGTCACGTAGGACTGTCAGAGACA................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................TTACGTGTTAGTCACGTAGGACTGTCAGA....................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TAGTCACGTAGGACTGTCAGAGACAcc................................................................................. | 27 | cc | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TGTTAGTCACGTAGGACTGTCAGAGACA................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .............................................................................................................................GTGTTAGTCACGTAGG................................................................................................ | 16 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - |
| CGGGCCCTACTCAACATCCACACCGATGTGGGCAGCGTGCGCATGCGGAGGCCTGTGCAGGGAGATGGGTCCTGGGGCGGGCACCCAGTGAGAACATAGGAGGTTTAGGGCCAACTGTTCATTACGTGTTAGTCACGTAGGACTGTCAGAGACAGCCCTTGTCCTGAGGCCCCTTTGTATTCCGCAGGGCAGAGAAGTCCCTGACCCCAGGACCACCTATAGCCTCTGGCAGCACAG ....................................................................................((.(((....))).))......(((((.....((((..((((((.....)))))))))).(((...))).))))).............................................................................. ...................................................................................84...........................................................................161.......................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................CGTAGGACTGTCAgaga......................................................................................... | 17 | gaga | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................GCCCTTGTCCTGAGGCCCCTTTGTA.......................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................GCACCCAGTGAGAACATAGGAGGTTTA.................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .............................................................................................................GCCAACTGTTCATTACGTGTTAGTCACG.................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................CTTGTCCTGAGGCCCCTTTGTATTCCG..................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |