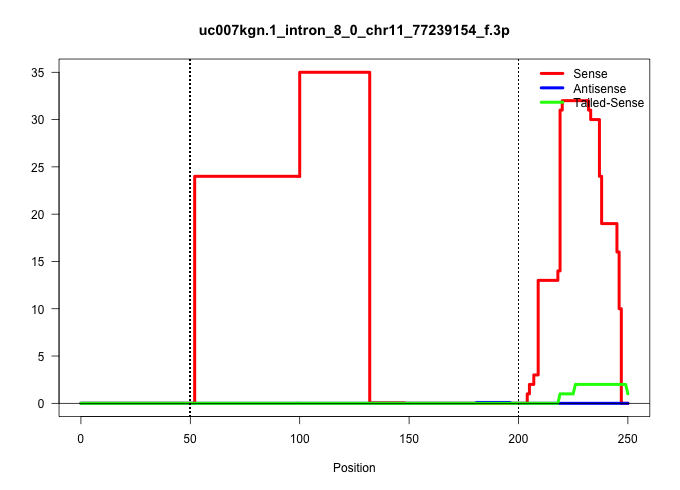
| Gene: mSSH-2L | ID: uc007kgn.1_intron_8_0_chr11_77239154_f.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(3) PIWI.ip |
(1) PIWI.mut |
(19) TESTES |
| TAATTTCTTGTACTATCATTAATAATACTAGATGTGACATTAATAATACTAGATGTGATAACATTAAGTTCCTGGTATTAAGAGGCATTGAGTACATGTTATTGAGTCACTGTTTAGAACCTCTGTTTCCCCCATGTTCCTTTTAGGATGCTGTGTTTGGTTGACTTGCTAATACTCACCTTGTCTTTTTTACTTCATAGGGCTCAGAATGGAATGCCTCGAACTTAGAGGACTTGCAGAACCGAGGGTG ......................................................................................................(((((......((((.......(((..((.((((((((.....))).)).)))...))..)))...)))).)))))........................................................................ ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................TGTGACATTAATAATACTAGATGTGATAACATTAAGTTCCTGGTATTAAGAG...................................................................................................................................................................... | 52 | 1 | 80.00 | 80.00 | 80.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................ATGTGATAACATTAAGTTCCTGGTATTAAGAGGCATTGAGTACATGTTATTG.................................................................................................................................................. | 52 | 1 | 24.00 | 24.00 | 24.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................CGAACTTAGAGGACTTGCAGAACCGAGG... | 28 | 1 | 10.00 | 10.00 | - | - | 3.00 | 3.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - |
| .........................................................................................................................................................................................................................CTCGAACTTAGAGGACTTGCAGAACCGAGG... | 30 | 1 | 7.00 | 7.00 | - | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................CTTAGAGGACTTGCAGAACC....... | 20 | 1 | 6.00 | 6.00 | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................TGGAATGCCTCGAACTTAGAGGACTTGC............. | 28 | 1 | 5.00 | 5.00 | - | - | - | 1.00 | 1.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................TGGAATGCCTCGAACTTAGAGGACTTGCA............ | 29 | 1 | 5.00 | 5.00 | - | - | - | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...........................................................................................................................................................................................................................CGAACTTAGAGGACTTGCAGAACCGAG.... | 27 | 1 | 5.00 | 5.00 | - | - | 3.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - |
| .................................................................................................................................................................................................................................TAGAGGACTTGCAGAACCGAGGGgt | 25 | gt | 5.00 | 0.00 | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................AACTTAGAGGACTTGCAGAACCGAGGGTG | 29 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................TAGAGGACTTGCAGAAC........ | 17 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................CGAACTTAGAGGACTTGCAGAACCGA..... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................CGAACTTAGAGGACTTGCAGAACCGAGGGgt | 31 | gt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................................................................................................................................................................AGAATGGAATGCCTCGAACTTAGAGGACTTGC............. | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................AATGGAATGCCTCGAACTTAGAGGA.................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................AGAGGACTTGCAGAACCGAGGGgta | 25 | gta | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................TCGAACTTAGAGGACTTGCAGAACCGAG.... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .........................................................................................................................................................................................................................CTCGAACTTAGAGGACTTGCA............ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................GAACTTAGAGGACTTGCAGAACCGA..... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................CAGAATGGAATGCCTCGAACTTAGAGGAC................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .....................................................................................................................................ATGTTCCTTTTAGGA...................................................................................................... | 15 | 19 | 0.05 | 0.05 | - | - | - | - | - | - | - | - | - | - | - | 0.05 | - | - | - | - | - | - | - |
| TAATTTCTTGTACTATCATTAATAATACTAGATGTGACATTAATAATACTAGATGTGATAACATTAAGTTCCTGGTATTAAGAGGCATTGAGTACATGTTATTGAGTCACTGTTTAGAACCTCTGTTTCCCCCATGTTCCTTTTAGGATGCTGTGTTTGGTTGACTTGCTAATACTCACCTTGTCTTTTTTACTTCATAGGGCTCAGAATGGAATGCCTCGAACTTAGAGGACTTGCAGAACCGAGGGTG ......................................................................................................(((((......((((.......(((..((.((((((((.....))).)).)))...))..)))...)))).)))))........................................................................ ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................................TGTCTTTTTTACTTCA..................................................... | 16 | 11 | 0.09 | 0.09 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.09 |