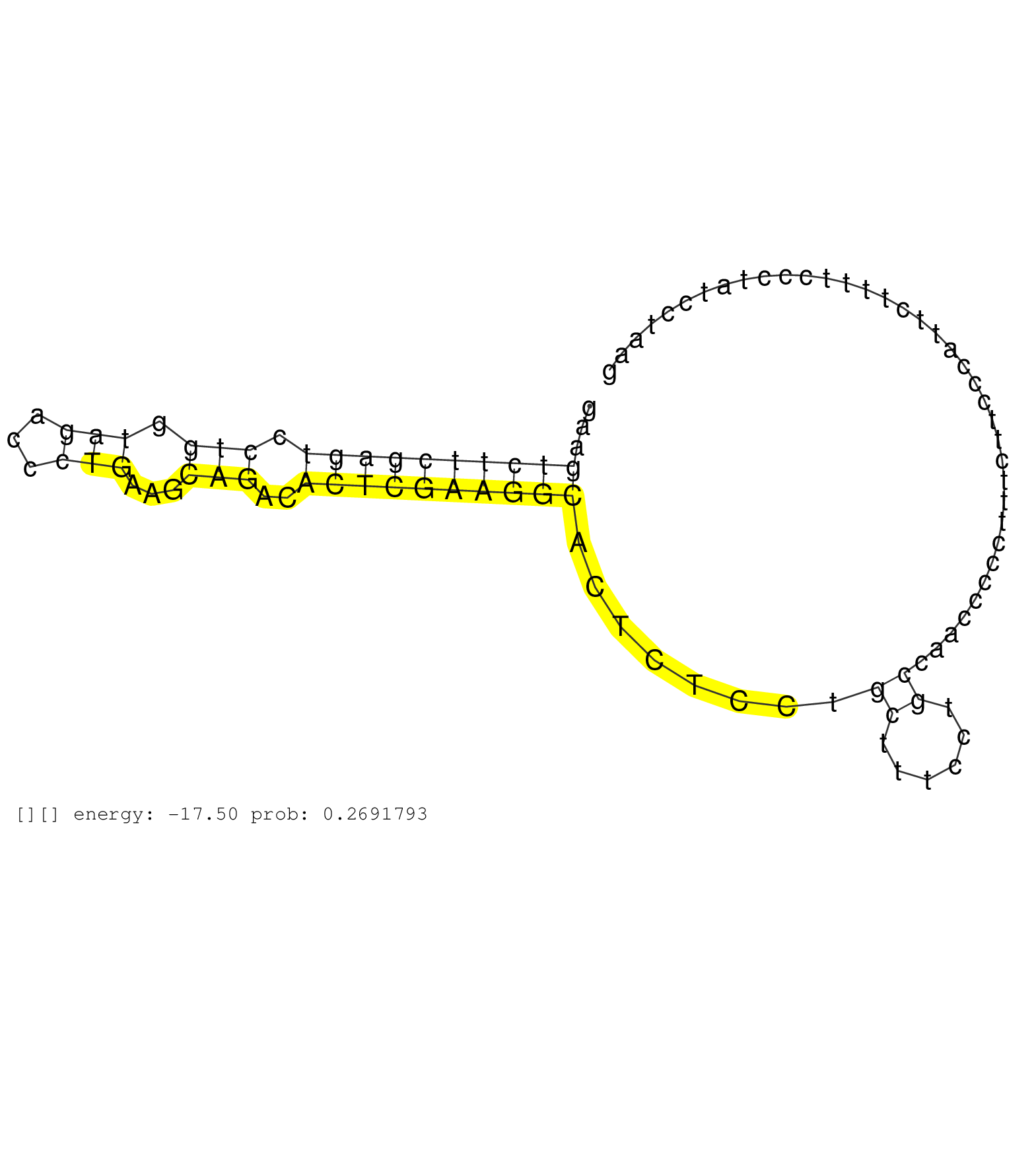
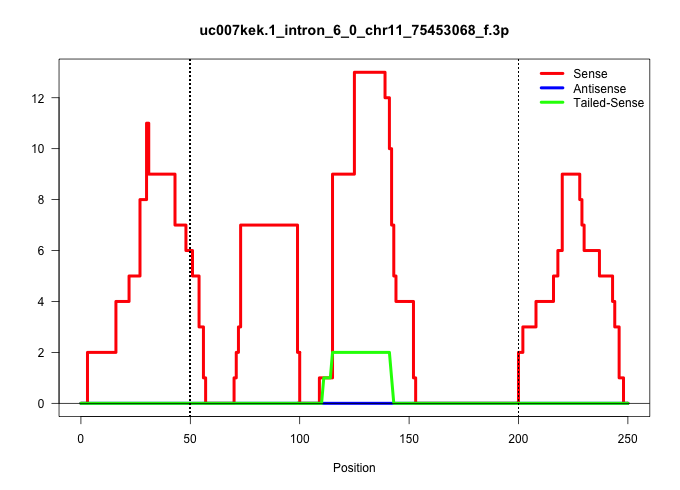
| Gene: Pps | ID: uc007kek.1_intron_6_0_chr11_75453068_f.3p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(14) TESTES |
| TCCTAAATCACAGCTTCATTCATAAACTCCTGGGCTTAGCAGAGGGTCTGCAGTTGATTGCTGCTGCTCCTGTTGGATACTTTGACCTGCCAGTGGACCAGAAGTCTTCGAGTCCTGGTAGACCCTGAAGCAGACACTCGAAGGCACTCTCCTGCTTTCCTGCCAACCCCCTTTCTTCCCATTCTTTTCCCTATCCTAAGTGAGAAAAAACGCAAGCCTGCGTGGACTGACCGCATCCTGTGGAGGTTGA .......................................................................................................((((((((((.(((.(((...)))...)))..))))))))))........((......))....................................................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................TGGTAGACCCTGAAGCAGACACTCGA............................................................................................................. | 26 | 1 | 3.00 | 3.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .........................................................................TGGATACTTTGACCTGCCAGTGGACC....................................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................................................................................TGAAGCAGACACTCGAAGGCACTCTCC.................................................................................................. | 27 | 1 | 3.00 | 3.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TGGTAGACCCTGAAGCAGACACTCGAA............................................................................................................ | 27 | 1 | 3.00 | 3.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................CGTGGACTGACCGCATCCTGTGGAGG.... | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................CATTCATAAACTCCTGGGCTTAGCAGA............................................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................TGGGCTTAGCAGAGGGTCTGCAGTTG.................................................................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .........................................................................TGGATACTTTGACCTGCCAGTGGACCA...................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...TAAATCACAGCTTCATTCATAAACTCCT........................................................................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| ...........................TCCTGGGCTTAGCAGAGGGTCTGCAGT.................................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TGGTAGACCCTGAAGCAGACACTCGAAG........................................................................................................... | 28 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................TCCTGGGCTTAGCAGAGGGTCTGC....................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..........................................................................................................................................................................................................AGAAAAAACGCAAGCCTGCGTGGACTGA.................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .......................................................................GTTGGATACTTTGACCTGCCAGTGGACC....................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................GTCCTGGTAGACCCTGAAGCAGACACTCGtt............................................................................................................ | 31 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................................................................................................................................................................................TGAGAAAAAACGCAAGCCTGCGTGGACT...................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................TTGGATACTTTGACCTGCCAGTGGACC....................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................TAAACTCCTGGGCTTAGCAGAGGGTC.......................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................................................................TGTTGGATACTTTGACCTGCCAGTGGACC....................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................TGAAGCAGACACTCGAAGGCACTCTCCT................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................TGAGAAAAAACGCAAGCCTGCGTGGACTG..................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................TGCGTGGACTGACCGCATCCTGTGGA...... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TGGTAGACCCTGAAGCAGACACTCGAAGG.......................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................GAGTCCTGGTAGACCCTGAAGCAGACACTC............................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................CCTGCGTGGACTGACCGCATCCTGTGG....... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................CGTGGACTGACCGCATCCTGTGGAGGTT.. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................AACGCAAGCCTGCGTGGACTGACCGCATC............. | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TGGTAGACCCTGAAGCAGACACTCGAAaa.......................................................................................................... | 29 | aa | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..............................TGGGCTTAGCAGAGGGTCTGCAGTTGA................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TGGTAGACCCTGAAGCAGACACTCGAAt........................................................................................................... | 28 | t | 1.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TCCTAAATCACAGCTTCATTCATAAACTCCTGGGCTTAGCAGAGGGTCTGCAGTTGATTGCTGCTGCTCCTGTTGGATACTTTGACCTGCCAGTGGACCAGAAGTCTTCGAGTCCTGGTAGACCCTGAAGCAGACACTCGAAGGCACTCTCCTGCTTTCCTGCCAACCCCCTTTCTTCCCATTCTTTTCCCTATCCTAAGTGAGAAAAAACGCAAGCCTGCGTGGACTGACCGCATCCTGTGGAGGTTGA .......................................................................................................((((((((((.(((.(((...)))...)))..))))))))))........((......))....................................................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|