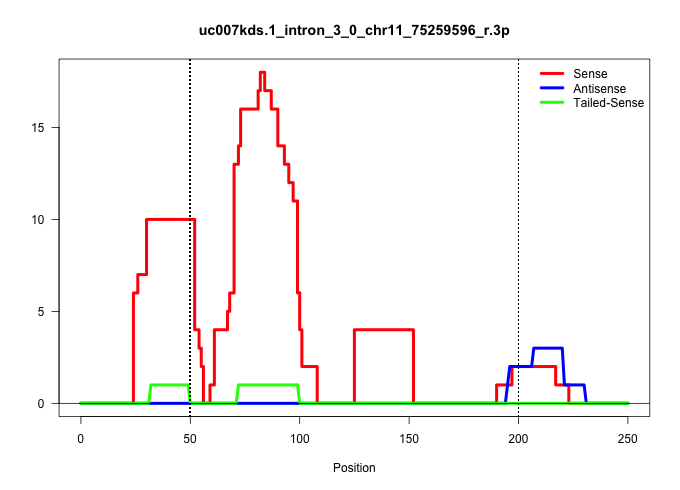
| Gene: Wdr81 | ID: uc007kds.1_intron_3_0_chr11_75259596_r.3p | SPECIES: mm9 |
 |
|
(2) OTHER.mut |
(4) PIWI.ip |
(3) PIWI.mut |
(16) TESTES |
| CTCTATAGAGACAGCACTCATGTCTACGTATCGATAAAGGAAGCTGTTTTGGGGGGAGGAAACAGGCTTACAGACGAGAACTTAATAACCCAGAGTAGCCCAGTCAGTACCTTGCCTTGGCTTCTGTTGTTTCTAAGCTCTGGGTAGATAGCTACCTTGCCATCTTCCCTGATCTTAGAACTTTCCCCACTCCCCTGTAGGTGACATCATCCGGAAAATCATCCCCAACCATGAGTTGGTCGGGGAGCTG |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................TACGTATCGATAAAGGAAGCTGTTTTGG...................................................................................................................................................................................................... | 28 | 1 | 6.00 | 6.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................CAGACGAGAACTTAATAACCCAGAGTAGC....................................................................................................................................................... | 29 | 1 | 5.00 | 5.00 | - | 3.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................GTTGTTTCTAAGCTCTGGGTAGATAGC.................................................................................................. | 27 | 1 | 4.00 | 4.00 | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................TCGATAAAGGAAGCTGTTTTGGGGGG.................................................................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................ACAGGCTTACAGACGAGAACTTAATAACC................................................................................................................................................................ | 29 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................................................................ACGAGAACTTAATAACCCAGAGTAGCCC..................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................ACAGGCTTACAGACGAGAACTTAATA................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................TTACAGACGAGAACTTAATAACCCAG............................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................CAGACGAGAACTTAATAACCCAGAGTA......................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................TAGGTGACATCATCCGGAAAATCATC........................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................TACAGACGAGAACTTAATAACCCAGAG........................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ......................................................................CAGACGAGAACTTAATAACCCAGAGTAGCC...................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................CGTATCGATAAAGGAAGCTGTTTTGGGG.................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............................................................................................................................................................................................TCCCCTGTAGGTGACATCATCCGGAAA................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................TAATAACCCAGAGTAGCCCAGTCAGT.............................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GACGAGAACTTAATAACCCAGAGTAGCC...................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................TCGATAAAGGAAGCTGTTTTGGGGG................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GACGAGAACTTAATAACCCAGAGTAGCt...................................................................................................................................................... | 28 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................................................................TTAATAACCCAGAGTAGCCCAGTCAGT.............................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ................................GATAAAGGAAGCTGattg........................................................................................................................................................................................................ | 18 | attg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...........................................................AAACAGGCTTACAGACGAGAACTTA...................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| CTCTATAGAGACAGCACTCATGTCTACGTATCGATAAAGGAAGCTGTTTTGGGGGGAGGAAACAGGCTTACAGACGAGAACTTAATAACCCAGAGTAGCCCAGTCAGTACCTTGCCTTGGCTTCTGTTGTTTCTAAGCTCTGGGTAGATAGCTACCTTGCCATCTTCCCTGATCTTAGAACTTTCCCCACTCCCCTGTAGGTGACATCATCCGGAAAATCATCCCCAACCATGAGTTGGTCGGGGAGCTG |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................................................CATCCGGAAAATCATCCCCAACCA................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................................................................................................................................................................................GAAAATCATCCCCggt........................ | 16 | ggt | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................TGTAGGTGACATCATCCGGAAAATCA............................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................GTAGGTGACATCATCCGGAAAATCA............................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |