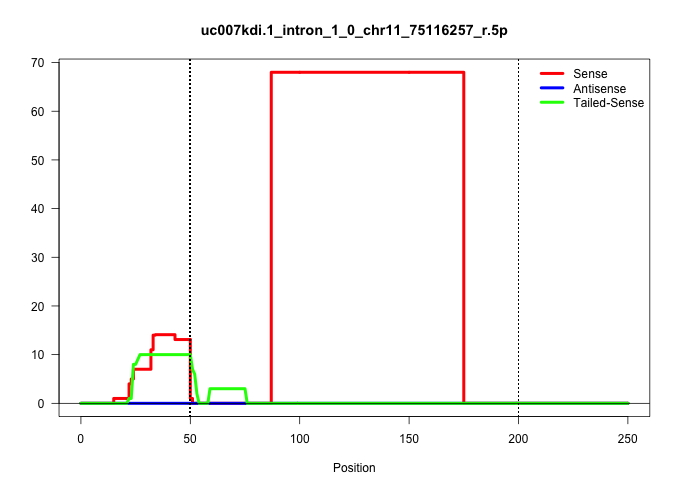
| Gene: Rpa1 | ID: uc007kdi.1_intron_1_0_chr11_75116257_r.5p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(3) PIWI.ip |
(1) PIWI.mut |
(17) TESTES |
| AGGCCATCCTTGGACAGAACACTATGTACCTTGGGGAACTCAAAGAGAAGGTTGGCTTTGTGTTTGTCCGCGCTGTGATAATGCTGGCCTTATGCCACTGAGCTGTTGAATGCTTGCTTTGGGTGTGGTGGATGCGCGCACTGAGTGAGGACTTACCTTTGTCTTTTCTGTATGTATCTGGTGTGAGGTAATGCGTCAGGGAATGCCATTGCACAGAAGCACAAGCCACCTTTGCCCCATCCCCATCGCT ...................................................................(((((((..........((((.....))))..((((........))))......))))))).(((((((..((..(((.((((.....))))...)))...))..)))))))....................................................................... ......................................................55..........................................................................................................................179..................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................CCTTATGCCACTGAGCTGTTGAATGCTTGCTTTGGGTGTGGTGGATGCGCGC............................................................................................................... | 52 | 1 | 75.00 | 75.00 | 75.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................GGGAACTCAAAGAGAAG........................................................................................................................................................................................................ | 17 | 1 | 5.00 | 5.00 | - | 1.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................TATGTACCTTGGGGAACTCAAAGAGAAG........................................................................................................................................................................................................ | 28 | 1 | 4.00 | 4.00 | - | - | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................GGGGAACTCAAAGAGAAG........................................................................................................................................................................................................ | 18 | 1 | 4.00 | 4.00 | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...........................................................GTGTTTGTCCGCGgagg.............................................................................................................................................................................. | 17 | gagg | 3.00 | 0.00 | - | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ........................TGTACCTTGGGGAACTCAAAGAGAAGaac..................................................................................................................................................................................................... | 29 | aac | 3.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - |
| ........................TGTACCTTGGGGAACTCAAAGAGAAGa....................................................................................................................................................................................................... | 27 | a | 2.00 | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........TGGACAGAACACTATGTACCTTGGGGA..................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....ATCCTTGGACAGAACACT................................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................ACCTTGGGGAACTCAAAGAGAAGa....................................................................................................................................................................................................... | 24 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ......................TATGTACCTTGGGGAACTCAAAGAGAA......................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................TGTACCTTGGGGAACTCAAAGAGAAGG....................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............AGAACACTATGTACCTTGGGGAACTCttc.............................................................................................................................................................................................................. | 29 | ttc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ......................TATGTACCTTGGGGAACTCAAAGAGAAGaac..................................................................................................................................................................................................... | 31 | aac | 1.00 | 4.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................TTGGGGAACTCAAAGAGAAGa....................................................................................................................................................................................................... | 21 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................ATGTACCTTGGGGAACTCAAAGAGAAG........................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................TGTACCTTGGGGAACTCAAAGAGAAGaacg.................................................................................................................................................................................................... | 30 | aacg | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................TACCTTGGGGAACTCAAAGAGAAGaacg.................................................................................................................................................................................................... | 28 | aacg | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............AGAACACTATGTACCTTGGGGAACTCAA............................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................TGTACCTTGGGGAACTCAAAGAGAAG........................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............CAGAACACTATGTACCTTGGGGAACT.................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................TGTACCTTGGGGAACTCAAAGAGAAGaa...................................................................................................................................................................................................... | 28 | aa | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................CTTGGGGAACTCAAAGAGAA......................................................................................................................................................................................................... | 20 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - |
| ..................................GGAACTCAAAGAGAAG........................................................................................................................................................................................................ | 16 | 10 | 0.10 | 0.10 | - | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGGCCATCCTTGGACAGAACACTATGTACCTTGGGGAACTCAAAGAGAAGGTTGGCTTTGTGTTTGTCCGCGCTGTGATAATGCTGGCCTTATGCCACTGAGCTGTTGAATGCTTGCTTTGGGTGTGGTGGATGCGCGCACTGAGTGAGGACTTACCTTTGTCTTTTCTGTATGTATCTGGTGTGAGGTAATGCGTCAGGGAATGCCATTGCACAGAAGCACAAGCCACCTTTGCCCCATCCCCATCGCT ...................................................................(((((((..........((((.....))))..((((........))))......))))))).(((((((..((..(((.((((.....))))...)))...))..)))))))....................................................................... ......................................................55..........................................................................................................................179..................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................ACTGAGCTGTTGAAattt............................................................................................................................................ | 18 | attt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |