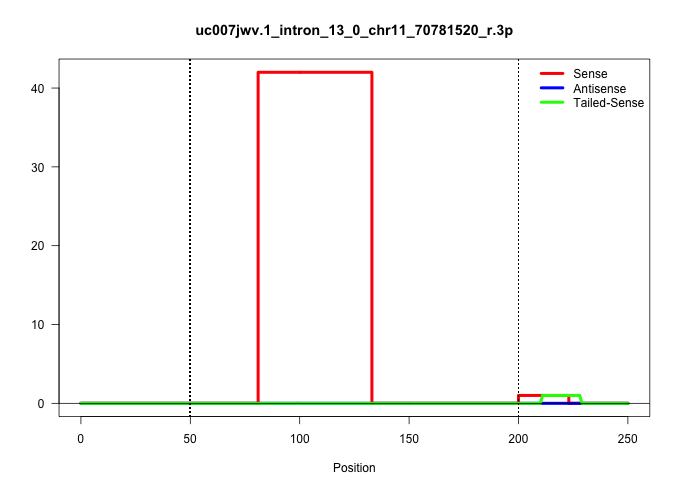
| Gene: Nup88 | ID: uc007jwv.1_intron_13_0_chr11_70781520_r.3p | SPECIES: mm9 |
 |
|
(1) PIWI.mut |
(4) TESTES |
| AGCAAGTGCTTTTAGGTGTTGTCATCTATCAAGAACCTAGAGATTATTTCTTGGGGAATAATCAGCTAATGACACAGTTAGTACATGCTCAGAATGCTATTTATAACTTTAGAAATTTAAAACGTGATTATGGTTTCAAGCAACTCTGAAATAAATTTATATTAAAATTAACTATTTTTACTAAATGGTTTTACTTTCAGAGATTACTCTGCATTAATCCACCCTTGTTTGAAATCCATCAAGTCTTGTT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) |
|---|---|---|---|---|---|---|---|---|
| .................................................................................TACATGCTCAGAATGCTATTTATAACTTTAGAAATTTAAAACGTGATTATGG..................................................................................................................... | 52 | 1 | 42.00 | 42.00 | 42.00 | - | - | - |
| ...................................................................................................................................................................................................................CATTAATCCACCCTcgga..................... | 18 | cgga | 1.00 | 0.00 | - | 1.00 | - | - |
| ........................................................................................................................................................................................................AGATTACTCTGCATTAATCCACC........................... | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 |
| AGCAAGTGCTTTTAGGTGTTGTCATCTATCAAGAACCTAGAGATTATTTCTTGGGGAATAATCAGCTAATGACACAGTTAGTACATGCTCAGAATGCTATTTATAACTTTAGAAATTTAAAACGTGATTATGGTTTCAAGCAACTCTGAAATAAATTTATATTAAAATTAACTATTTTTACTAAATGGTTTTACTTTCAGAGATTACTCTGCATTAATCCACCCTTGTTTGAAATCCATCAAGTCTTGTT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) |
|---|---|---|---|---|---|---|---|---|
| ...........................................................................TTAGTACATGCTCAGAAag............................................................................................................................................................ | 19 | ag | 1.00 | 0.00 | - | - | 1.00 | - |
| .........................................................AATCAGCTAATGACACgt............................................................................................................................................................................... | 18 | gt | 1.00 | 0.00 | - | 1.00 | - | - |