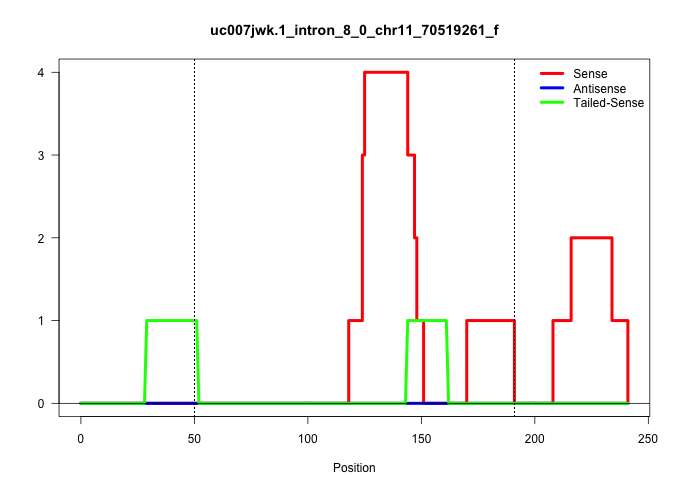
| Gene: Kif1c | ID: uc007jwk.1_intron_8_0_chr11_70519261_f | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(2) PIWI.ip |
(1) PIWI.mut |
(10) TESTES |
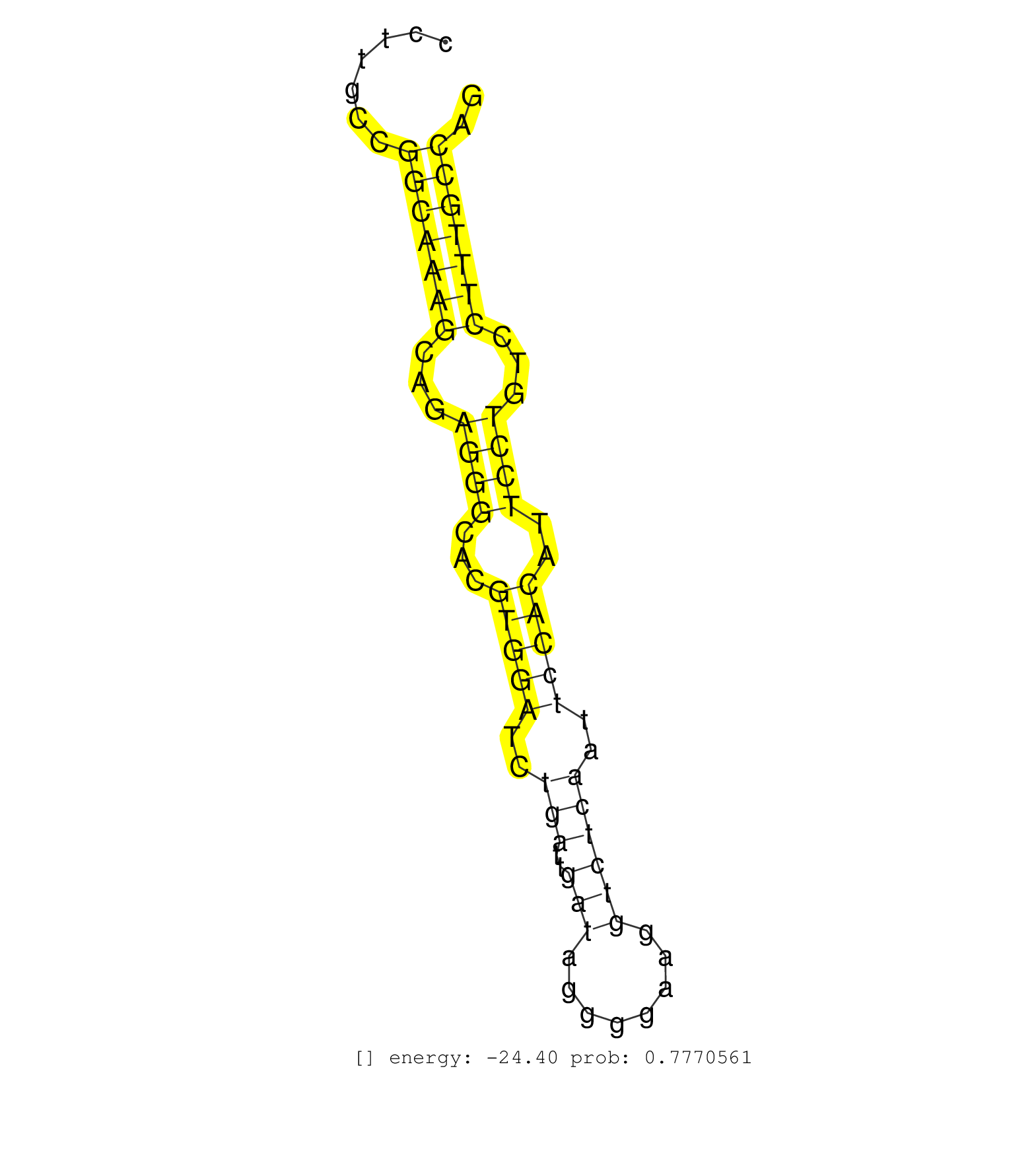
| CCGGGAGTGAGCGGGCTGACTCCTCAGGGGCCCGGGGGATGCGTCTAAAGGTGAGGATGGTGGATGCCTCAGGGGCTGTGCAGGGAGGGAGGTTGTTGATCGTCCCCTAGCACCCTTGCCGGCAAAGCAGAGGGCACGTGGATCTGATTGATAGGGGAAGGTCTCAATTCCACATTCCTGTCCTTTGCCAGGAAGGCGCCAACATCAATAAGTCCCTGACTACGCTTGGGAAGGTGATCTC ........................................................................................................................(((((((...((((...(((((..(((..(((........))))))..)))))..))))...))))))).................................................... .................................................................................................................114..........................................................................191................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesKO6() Testes Data. (Zcchc11 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037902(GSM510438) testes_rep3. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................GAGGGCACGTGGATCTGAa............................................................................................. | 19 | a | 4.00 | 0.00 | 4.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................CCGGCAAAGCAGAGGGCACGTGGATC................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - |
| ............................................................................................................................AAGCAGAGGGCACGTGGATCTGA.............................................................................................. | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................GCCCGGGGGATGCGTCTAAAGGa............................................................................................................................................................................................. | 23 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - |
| ..........................................................................................................................................................................CACATTCCTGTCCTTTGCCAG.................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 |
| ............................................................................................................................AAGCAGAGGGCACGTGGATCTGAT............................................................................................. | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGACTACGCTTGGGAAGGTGATCTC | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - |
| ................................................................................................................................................TGATTGATAGGGGActta............................................................................... | 18 | ctta | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................................................................................AGCAGAGGGCACGTGGATCTGATTGA.......................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ................................................................................................................................................................................................................TAAGTCCCTGACTACGCTTGGGAAGG....... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| CCGGGAGTGAGCGGGCTGACTCCTCAGGGGCCCGGGGGATGCGTCTAAAGGTGAGGATGGTGGATGCCTCAGGGGCTGTGCAGGGAGGGAGGTTGTTGATCGTCCCCTAGCACCCTTGCCGGCAAAGCAGAGGGCACGTGGATCTGATTGATAGGGGAAGGTCTCAATTCCACATTCCTGTCCTTTGCCAGGAAGGCGCCAACATCAATAAGTCCCTGACTACGCTTGGGAAGGTGATCTC ........................................................................................................................(((((((...((((...(((((..(((..(((........))))))..)))))..))))...))))))).................................................... .................................................................................................................114..........................................................................191................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesKO6() Testes Data. (Zcchc11 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037902(GSM510438) testes_rep3. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................GAGGATGGTGGATGCgtgt.............................................................................................................................................................................. | 19 | gtgt | 2.00 | 0.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - |
| .................................................GAGGATGGTGGATGCgtg.............................................................................................................................................................................. | 18 | gtg | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - |