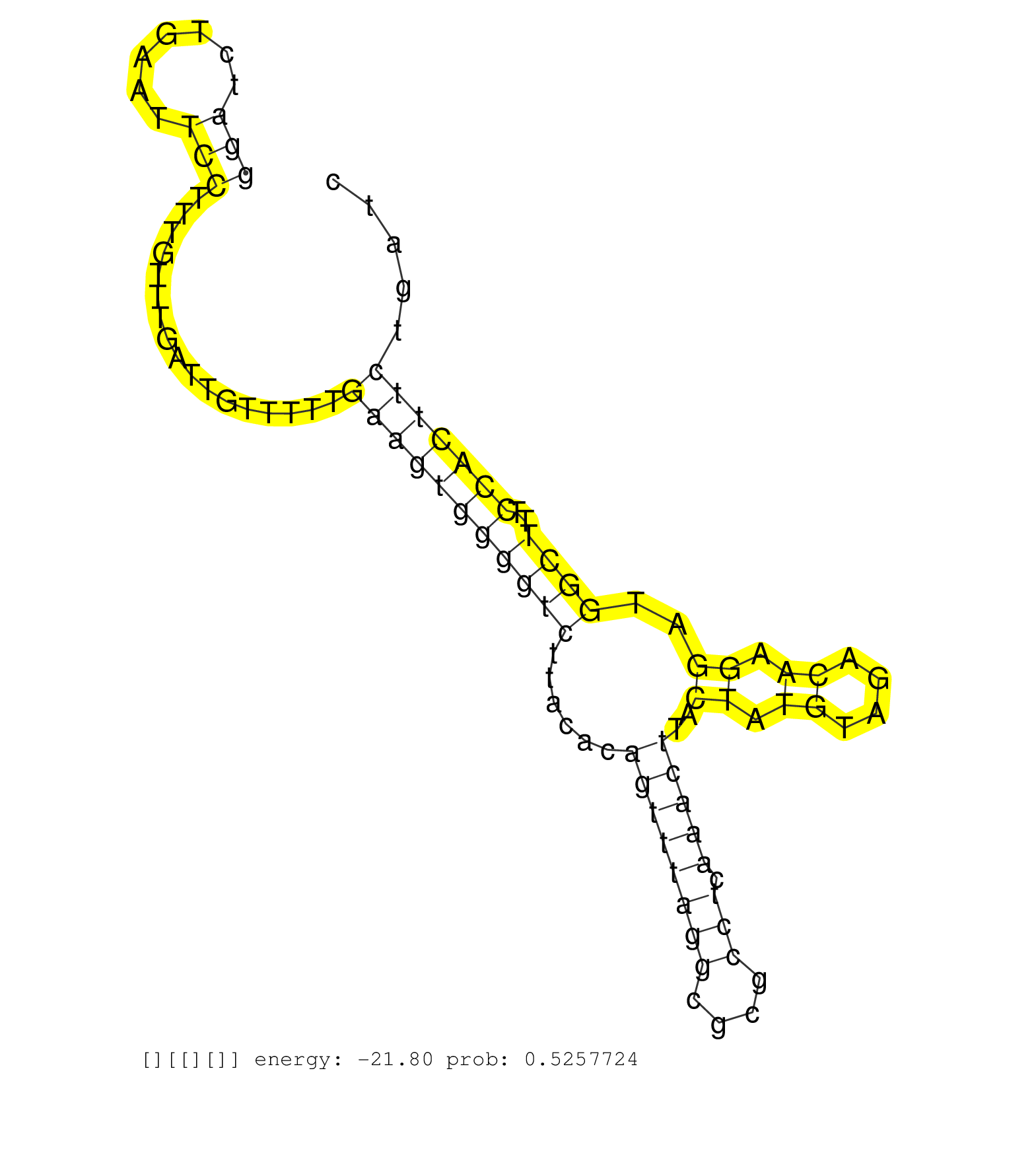

| Gene: AI842396 | ID: uc007jwg.1_intron_4_0_chr11_70504055_r.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(16) TESTES |
| TTTTCTCAGAGTAGAAGCTAGCAAGTCCCAGCAATTCAGCCCCTTCCAAGGTAGGGCTACAAATGTGTGGTCACTTAGCTTGTTAGATGGGTACTGGGATCTGAATTCCTTTGTTTGATTGTTTTTGAAGTGGGGTCTTACACAGTTTAGGCGCGCCTCAAACTTACTATGTAGACAAGGATGGCTTTCCACTTCTGATCCTGTCATCATCCCCAAGTGCTTAGATTACACACATGTACCAGGGGGTGTT ................................................................................................(((.......))).................(((((((((((......((((((((....))).)))))..((.((....)).))..))))..)))))))....................................................... ................................................................................................97.....................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................TACTATGTAGACAAGGATGGCTTTCCAC.......................................................... | 28 | 1 | 8.00 | 8.00 | 4.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ....................................................................................................................................................................TACTATGTAGACAAGGATGGCTTTCCACT......................................................... | 29 | 1 | 6.00 | 6.00 | 1.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - |
| ....................................................................................................................................................................TACTATGTAGACAAGGATGGCTTTCC............................................................ | 26 | 1 | 3.00 | 3.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................CTTACTATGTAGACAAGGATGGCTTTCC............................................................ | 28 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..TTCTCAGAGTAGAAGCTAGCAAGTCCCAG........................................................................................................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................TTACTATGTAGACAAGGATGGCTTTCCACT......................................................... | 30 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TCTCAGAGTAGAAGCTAGCAAGTCCCAG........................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................................................................TACTATGTAGACAAGGATGGCTTTCCA........................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................ACTTACTATGTAGACAAGGATGGCTTTCC............................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......AGAGTAGAAGCTAGCAAGTCCCAGCAAT....................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................TGAATTCCTTTGTTTGATTGTTTTTG........................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................GCTAGCAAGTCCCAcgac........................................................................................................................................................................................................................ | 18 | cgac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .........................................................TACAAATGTGTGGTgtgg............................................................................................................................................................................... | 18 | gtgg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...................................................................................................................................................................TTACTATGTAGACAAGGATGGCTTTCC............................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TGATTGTTTTTGAAGTGGGGTCTTACA............................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TTAGATTACACACATGTACCAGGGGGTG.. | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TAGGCGCGCCTCAAACTTACTATGTAGA........................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................TTGTTTGATTGTTTTTGAAGTGGGG................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................TTACTATGTAGACAAGGATGGCTTTC............................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TTAGCTTGTTAGATGGGTACTGGGATC..................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................TTACTATGTAGACAAGGATGGCTTTCCAC.......................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................TACTATGTAGACAAGGATGGCTTTCCACTT........................................................ | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................TGTAGACAAGGATGGCTTTCCACTTC....................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| TTTTCTCAGAGTAGAAGCTAGCAAGTCCCAGCAATTCAGCCCCTTCCAAGGTAGGGCTACAAATGTGTGGTCACTTAGCTTGTTAGATGGGTACTGGGATCTGAATTCCTTTGTTTGATTGTTTTTGAAGTGGGGTCTTACACAGTTTAGGCGCGCCTCAAACTTACTATGTAGACAAGGATGGCTTTCCACTTCTGATCCTGTCATCATCCCCAAGTGCTTAGATTACACACATGTACCAGGGGGTGTT ................................................................................................(((.......))).................(((((((((((......((((((((....))).)))))..((.((....)).))..))))..)))))))....................................................... ................................................................................................97.....................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................CCTGTCATCATCCCCAAGTGCTTAGA......................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |