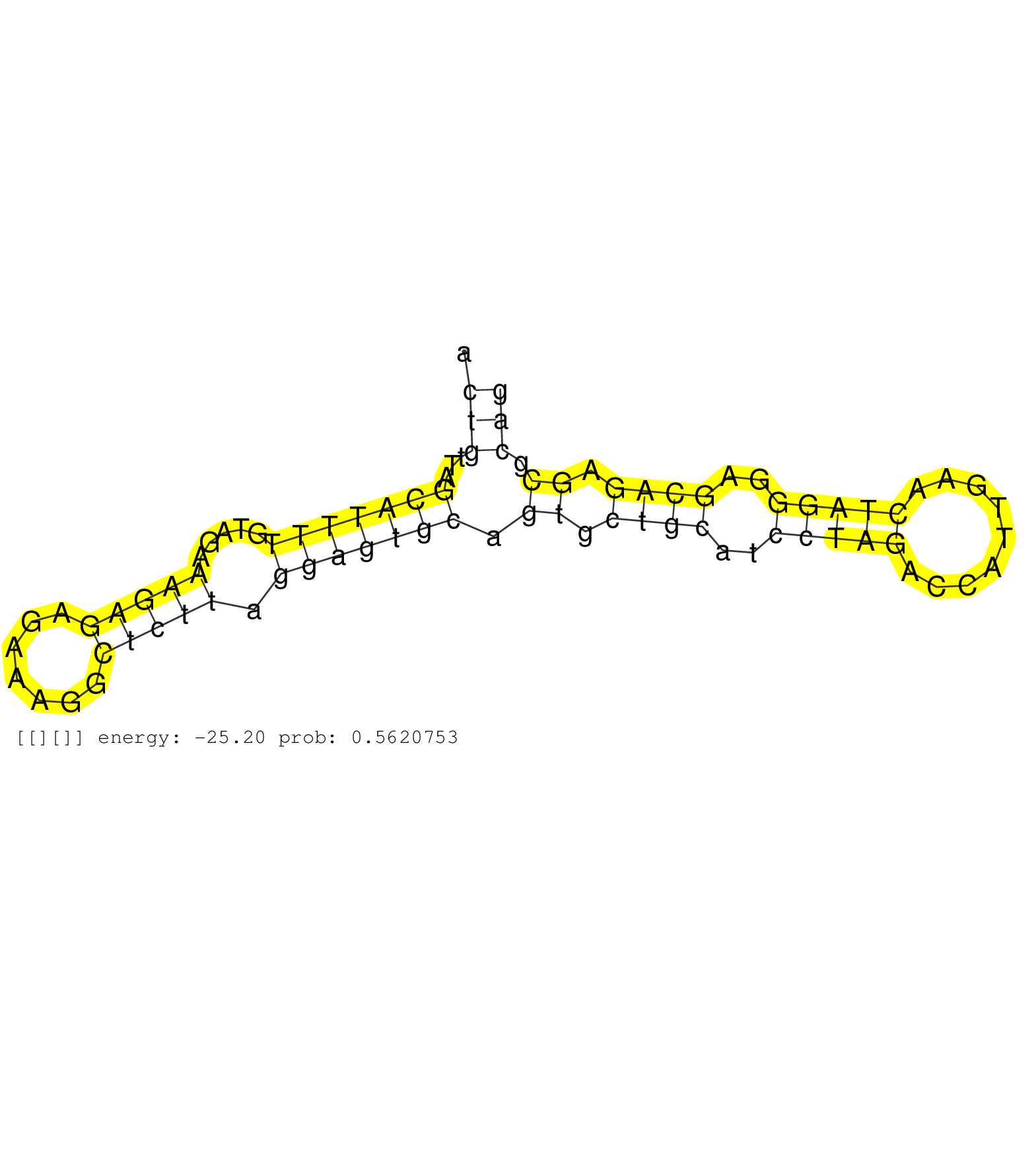

| Gene: Eno3 | ID: uc007jvx.1_intron_5_0_chr11_70472664_f.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(3) OTHER.mut |
(5) PIWI.ip |
(1) TDRD1.ip |
(21) TESTES |
| ATTTGATTTCCCTTAACTGTCAGAGCAGTAGGGAAGTAAGCACTGTTAGCATTTTGTAGAAAGAGAGAAAGGCTCTTAGGAGTGCAGTGCTGCATCCTAGACCATTGAACTAGGGAGCAGAGCGCAGCTGTCACCTCCAGTTCCCTCTCACCTTTGACCCCTATGCCACCCAGCTCTGAGCACCAACTTCTGTGTCCCAGGCCTTTAATGTGATCAACGGCGGCTCTCATGCTGGAAACAAGCTGGCCAT ..........................................(((...(((((((.....(((((.......))))).))))))).((.((((..(((((.........)))))..)))).)).)))........................................................................................................................... .........................................42...................................................................................127......................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | mjTestesWT2() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................TAGACCATTGAACTAGGGAGCAGAGC............................................................................................................................... | 26 | 1 | 10.00 | 10.00 | 4.00 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..............................................TAGCATTTTGTAGAAAGAGAGAAAGGC................................................................................................................................................................................. | 27 | 1 | 9.00 | 9.00 | 1.00 | 5.00 | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TAGACCATTGAACTAGGGAGCAGAGCG.............................................................................................................................. | 27 | 1 | 7.00 | 7.00 | 1.00 | - | - | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..............................................TAGCATTTTGTAGAAAGAGAGAAAGG.................................................................................................................................................................................. | 26 | 1 | 6.00 | 6.00 | 3.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TAGACCATTGAACTAGGGAGCAGAGCGC............................................................................................................................. | 28 | 1 | 5.00 | 5.00 | - | 1.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................TTGAACTAGGGAGCAGAGC............................................................................................................................... | 19 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............TAACTGTCAGAGCAGTAGGGAAGTAAGC................................................................................................................................................................................................................. | 28 | 1 | 4.00 | 4.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................TAGCATTTTGTAGAAAGAGAGAAAGGCTC............................................................................................................................................................................... | 29 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............TTAACTGTCAGAGCAGTAGGGAAGTAAGC................................................................................................................................................................................................................. | 29 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TGAACTAGGGAGCAGAGCGCAGCTGTC...................................................................................................................... | 27 | 1 | 3.00 | 3.00 | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................TAGCATTTTGTAGAAAGAGAGAAAGGCT................................................................................................................................................................................ | 28 | 1 | 3.00 | 3.00 | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................GACCATTGAACTAGGGAGCAGAGCGC............................................................................................................................. | 26 | 1 | 3.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TGAACTAGGGAGCAGAGCGCAGCTGT....................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................TGTCAGAGCAGTAGGGAAGTAAGCACT.............................................................................................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............TTAACTGTCAGAGCAGTAGGGAAGTAA................................................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................AATGTGATCAACGGCGGCTCTCAT.................... | 24 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................TTGAACTAGGGAGCAGAGCGCAGCTG........................................................................................................................ | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................TAGAAAGAGAGAAAGGCTCTTAGGAGT....................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................AGTGCAGTGCTGCATCtct....................................................................................................................................................... | 19 | tct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .................................................................................GTGCAGTGCTGCATCCctgg..................................................................................................................................................... | 20 | ctgg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ................................................................................................................................................................................................................................TCTCATGCTGGAAACAAGCTGGCCAT | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TTAATGTGATCAACGGCGGCTCcc...................... | 24 | cc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................TGTAGAAAGAGAGAAAGGCTCTTAGG.......................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .............................................................................................................................................................................................................TAATGTGATCAACGGCGGCTCTCATGCTG................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................................TTGAACTAGGGAGCAGAGCGCAGCTGT....................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGAGCACCAACTTCTGTGTCCCAGGa................................................ | 26 | a | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............TTAACTGTCAGAGCAGTAGGGAAGTAAGt................................................................................................................................................................................................................. | 29 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................GTAAGCACTGTTAGCATTTTGTAGAAA............................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...............................................AGCATTTTGTAGAAAGAGAGAAAGGC................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TAATGTGATCAACGGCGGCTCTCATG................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .................................................................................................TAGACCATTGAACTAGGGAGCAGAGt............................................................................................................................... | 26 | t | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................CATTGAACTAGGGAGCAGAGCGCAGC.......................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TAGACCATTGAACTAGGGAGCAGAG................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TTAATGTGATCAACGGCGGC.......................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................ATTTTGTAGAAAGAGAGAAAGGCTCTTAGGt......................................................................................................................................................................... | 31 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............TAACTGTCAGAGCAGTAGGGAAGTAAG.................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ATTTGATTTCCCTTAACTGTCAGAGCAGTAGGGAAGTAAGCACTGTTAGCATTTTGTAGAAAGAGAGAAAGGCTCTTAGGAGTGCAGTGCTGCATCCTAGACCATTGAACTAGGGAGCAGAGCGCAGCTGTCACCTCCAGTTCCCTCTCACCTTTGACCCCTATGCCACCCAGCTCTGAGCACCAACTTCTGTGTCCCAGGCCTTTAATGTGATCAACGGCGGCTCTCATGCTGGAAACAAGCTGGCCAT ..........................................(((...(((((((.....(((((.......))))).))))))).((.((((..(((((.........)))))..)))).)).)))........................................................................................................................... .........................................42...................................................................................127......................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | mjTestesWT2() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|