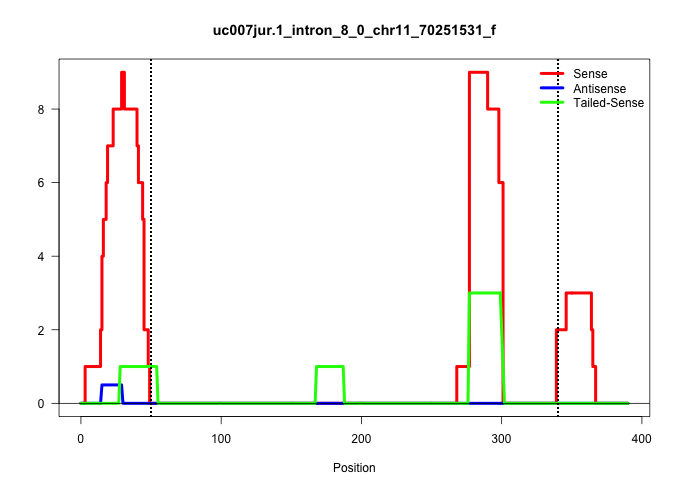
| Gene: Arrb2 | ID: uc007jur.1_intron_8_0_chr11_70251531_f | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(1) OVARY |
(3) PIWI.ip |
(1) PIWI.mut |
(15) TESTES |
| CCATGTCACCAACAATTCTGCCAAGACCGTCAAGAAGATCAGAGTGTCTGGTAGGAGGTGGGGGTGGGCATGGGGATAGGGAAGGAAGTCCCTACTTAGAAAGGCTGCTGCTGCCATGTCAGCCCAGGCCGTGTCGCAGTGTGGGAAACCTACCGGGCCATAAAGAATTGAGTCTGTTCCAGAAGAGATACATGGCCCTAGTGTTTACTATAGTAGGTCTGTTGGCCAGGGGTGGAAGGTGGAGGTGTGCTGCCCCCATGGGGTTTCCACCATGTGGTCGGGGGTTGTGGTGCCGAATTGTCTGTGTCACCCAAAGCCACTTGGTCCCCTGCCCCCACAGTGAGACAGTACGCCGACATTTGCCTCTTCAGCACCGCGCAGTACAAGTGT ...................................................................................................................................................................(((.....((((.((....)).)))).((..(((((....(((((....)))))......(((((..((((((((((((.(((.....))).))))....))))))))...))))).)))))..))...........)))....................................................................................... ...................................................................................................................................................................164..........................................................................................................................................305................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................................................................................................................................TCGGGGGTTGTGGTGCCGAATTGT......................................................................................... | 24 | 1 | 6.00 | 6.00 | 4.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .....................................................................................................................................................................................................................................................................................TCGGGGGTTGTGGTGCCGAAT............................................................................................ | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - |
| ..................................................................................................................................................................................................................TAGTAGGTCTGTTGGCCAGGGGTGG........................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............TTCTGCCAAGACCGTCAAGAAGATCA............................................................................................................................................................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................GCCAAGACCGTCAAGAAGATCAGAG.......................................................................................................................................................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................TGCCAAGACCGTCAAGAAGATCAGAGT......................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................................................................................................................................GTGAGACAGTACGCCGACATTTGCCT......................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................TGAGTCTGTTCCAGAAGgcc.......................................................................................................................................................................................................... | 20 | gcc | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................................................................TCGGGGGTTGTGGTGCCGAATTGa......................................................................................... | 24 | a | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................................................................TCGGGGGTTGTGGTGCCGActtg.......................................................................................... | 23 | cttg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...............TTCTGCCAAGACCGTCAAGAAGATCAGAGT......................................................................................................................................................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................................................................................................................................GTGAGACAGTACGCCGACATTTGCC.......................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............TTCTGCCAAGACCGTCAAGAAcacc.............................................................................................................................................................................................................................................................................................................................................................. | 25 | cacc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..............................................................................................................................................................................................................................................................................................................................ACTTGGTCCCCTGCCCCCAaa................................................... | 21 | aa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...TGTCACCAACAATTCTGCCAAGACCGTC....................................................................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................AGACCGTCAAGAAGATCAGAGTGTCT..................................................................................................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................GTCAAGAAGATCAGAGTGTCTGGggga............................................................................................................................................................................................................................................................................................................................................... | 27 | ggga | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................................................ACCATGTGGTCGGGGGTTGTGG.................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................................................................TCGGGGGTTGTGGTGCCGAATTatt........................................................................................ | 25 | att | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................TCAAGAAGATCAGAGTGTC...................................................................................................................................................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................TCTGCCAAGACCGTCAAGAAGATCAGAGT......................................................................................................................................................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................................................................................................................................................AGTACGCCGACATTTGCCTCT....................... | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............ATTCTGCCAAGACCGTCAAGAAGATC.............................................................................................................................................................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| CCATGTCACCAACAATTCTGCCAAGACCGTCAAGAAGATCAGAGTGTCTGGTAGGAGGTGGGGGTGGGCATGGGGATAGGGAAGGAAGTCCCTACTTAGAAAGGCTGCTGCTGCCATGTCAGCCCAGGCCGTGTCGCAGTGTGGGAAACCTACCGGGCCATAAAGAATTGAGTCTGTTCCAGAAGAGATACATGGCCCTAGTGTTTACTATAGTAGGTCTGTTGGCCAGGGGTGGAAGGTGGAGGTGTGCTGCCCCCATGGGGTTTCCACCATGTGGTCGGGGGTTGTGGTGCCGAATTGTCTGTGTCACCCAAAGCCACTTGGTCCCCTGCCCCCACAGTGAGACAGTACGCCGACATTTGCCTCTTCAGCACCGCGCAGTACAAGTGT ...................................................................................................................................................................(((.....((((.((....)).)))).((..(((((....(((((....)))))......(((((..((((((((((((.(((.....))).))))....))))))))...))))).)))))..))...........)))....................................................................................... ...................................................................................................................................................................164..........................................................................................................................................305................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............ATTCTGCCAAGACCG......................................................................................................................................................................................................................................................................................................................................................................... | 15 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| ...........................................................................................................................................................................................................................................................................................................................CTTGGTCCCCTGCCagta......................................................... | 18 | agta | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............TTCTGCCAAGACCGT........................................................................................................................................................................................................................................................................................................................................................................ | 15 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - |