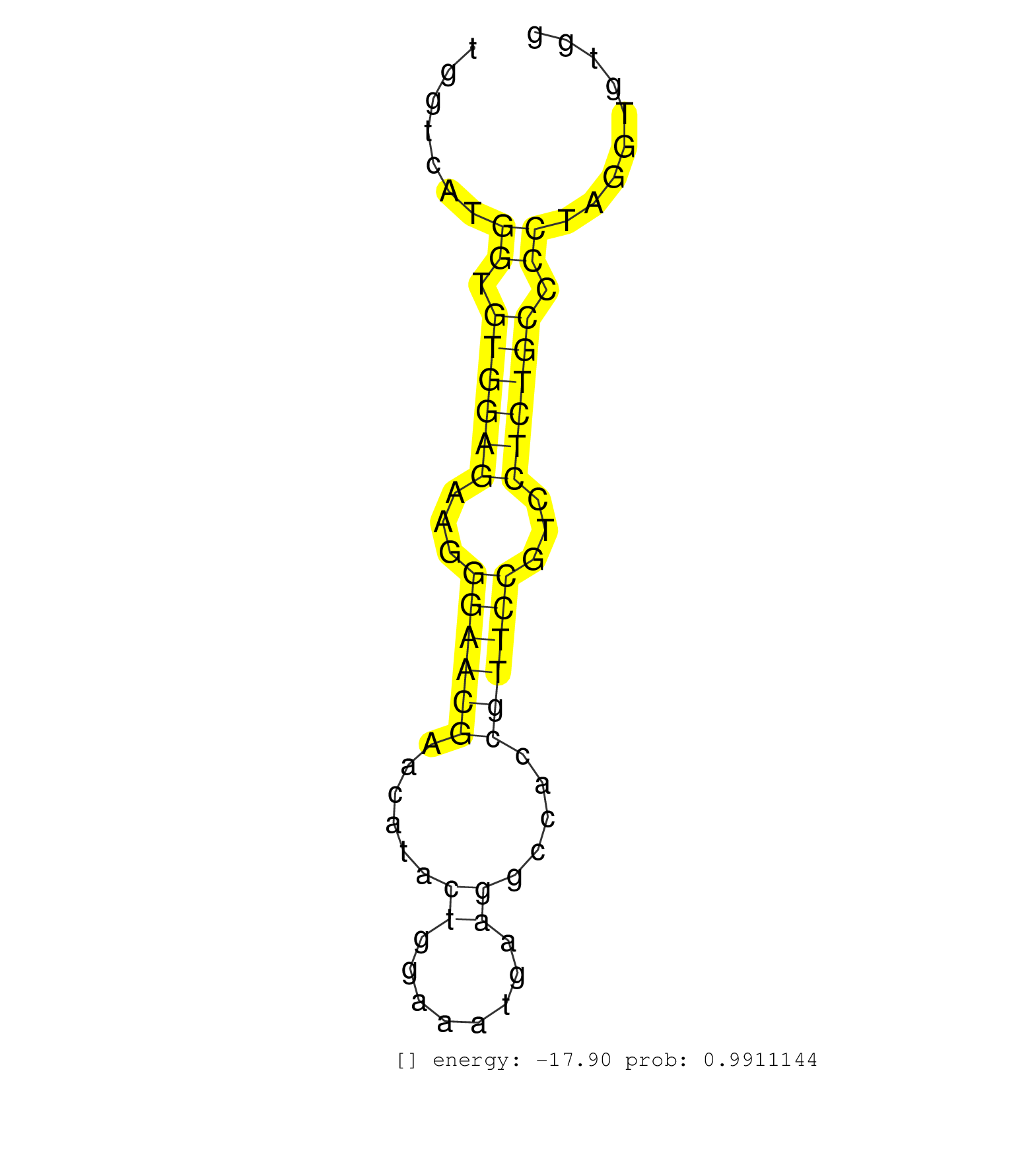

| Gene: Pelp1 | ID: uc007jup.1_intron_6_0_chr11_70210437_r | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(1) OVARY |
(4) PIWI.ip |
(3) PIWI.mut |
(28) TESTES |
| CCTTCTCTGCACCTTGAGGCCTTGGACTTGCTCTCTGCCCTGATCCTTGCGTAAGTAAACTTGGTCATGGTGTGGAGAAGGGAACGAACATACTGGAAATGAAGGCCACCGTTCCGTCCTCTGCCCCTAGGTGTGGAAGTCGACTTTTGCGTTTTGGTGCCCTGATCAGCCGGCTGCTTC ....................................................................((.((((((...((((((......((........)).....))))))...)))))).))..................................................... .............................................................62........................................................................136.......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR042485(GSM539877) mouse testicular tissue [09-002]. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................CATGGTGTGGAGAAGGGAACGA............................................................................................. | 22 | 1 | 11.00 | 11.00 | - | 4.00 | 4.00 | - | - | - | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................ATGGTGTGGAGAAGGGAACGA............................................................................................. | 21 | 1 | 11.00 | 11.00 | - | 1.00 | - | 2.00 | - | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - |
| ................................................................TCATGGTGTGGAGAAGGGAAC............................................................................................... | 21 | 1 | 7.00 | 7.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..................................................................ATGGTGTGGAGAAGGGAACGAA............................................................................................ | 22 | 1 | 3.00 | 3.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................ATGGTGTGGAGAAGGGAACG.............................................................................................. | 20 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................TTGGACTTGCTCTCTGCCC............................................................................................................................................ | 19 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTAAACTTGGTCATGGTGTGG......................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TGCGTTTTGGTGCCCTGATCAGCCGG....... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| ................................................................TCATGGTGTGGAGAAGGGAACG.............................................................................................. | 22 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .....TCTGCACCTTGAGGCCTTGGACTTGC..................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................ATGGTGTGGAGAAGGGAACGAAa........................................................................................... | 23 | a | 2.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TTCCGTCCTCTGCCCCTAGat................................................ | 21 | at | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................ATGGTGTGGAGAAGGGAACGAAga.......................................................................................... | 24 | ga | 1.00 | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................TCATGGTGTGGAGAAGGGAACGt............................................................................................. | 23 | t | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................TAAACTTGGTCATGGTGTGGAGAAGG................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................GAACGAACATACTGGAAATGAAGGCCAC....................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................AGGGAACGAACATACTGGAAATGAAGGC.......................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................GCGTTTTGGTGCCCTGATCAGCCGGCTt.... | 28 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................CATGGTGTGGAGAAGGGAAC............................................................................................... | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................TTGGACTTGCTCTCTGC.............................................................................................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................TCATGGTGTGGAGAAGGGAACGA............................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................CATGGTGTGGAGAAGGGAACG.............................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................TCATGGTGTGGAGAAGGGAACGAAaa.......................................................................................... | 26 | aa | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................CATGGTGTGGAGAAGGGAACGAACATA........................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................AACTTGGTCATGGTGTGGAGAAGGGAA................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTAAACTTGGTCATGGTGTG.......................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................GTTTTGGTGCCCTGATCAGCCGGCTGCT.. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TCTGCACCTTGAGGCCTTGGACTTG...................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................AACGAACATACTGGAAATGAAGGCCACC...................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CGACTTTTGCGTTTTGGTGCCCTGAT.............. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ......................................................................TGTGGAGAAGGGAACGAACATACTGGAA.................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TGGAAGTCGACTTTTGCGTTTTGGTGt.................... | 27 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................................................................................................................................................GTGCCCTGATCAGCCGGCTG.... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TGCGTTTTGGTGCCCTGATCAGCCGGC...... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............................................................................................................................................CGACTTTTGCGTTTa......................... | 15 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TGCGTTTTGGTGCCCTGATCAGCCGGCT..... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................TGGTGTGGAGAAGGGAACGA............................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTAAACTTGGTCATGGTGTGGAGA...................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................TGGTGTGGAGAAGGGAACGAACATACT...................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................ATGGTGTGGAGAAGGGA................................................................................................. | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................ATGGTGTGGAGAAGGG.................................................................................................. | 16 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - |
| CCTTCTCTGCACCTTGAGGCCTTGGACTTGCTCTCTGCCCTGATCCTTGCGTAAGTAAACTTGGTCATGGTGTGGAGAAGGGAACGAACATACTGGAAATGAAGGCCACCGTTCCGTCCTCTGCCCCTAGGTGTGGAAGTCGACTTTTGCGTTTTGGTGCCCTGATCAGCCGGCTGCTTC ....................................................................((.((((((...((((((......((........)).....))))))...)))))).))..................................................... .............................................................62........................................................................136.......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR042485(GSM539877) mouse testicular tissue [09-002]. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|