
| Gene: Tmem107 | ID: uc007jpc.1_intron_2_0_chr11_68884977_f.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(4) PIWI.ip |
(7) TESTES |
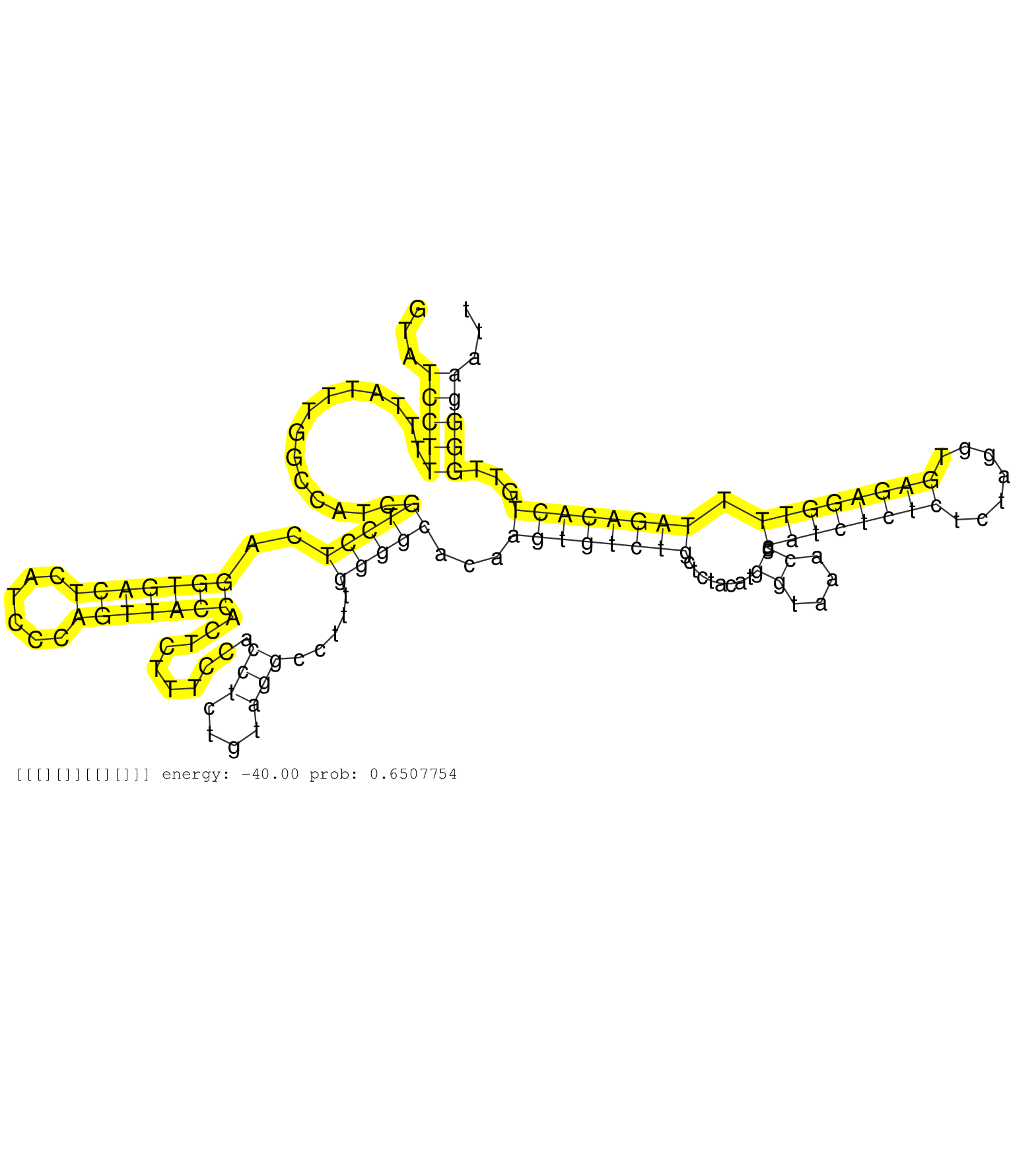
| TGGCTTCCTCTCAGGAGTCTCCATGTTCAATAGCACCCAGAGCCTCCTCTGTATCCTTTTTATTTGGCCATCGTCCTCAGGTGACTCATCCCAGTTACCACTCTTTCCACCTCTGTAGGCCTTTGGGGCACAAGTGTCTGCTCTACATGGTAAACCGAATCTCTCTCTAGGTGAGAGGTTTTAGACACTGTTGGGGAATTAATAGACTTCAAACATTCGTTAAATCCGGATGTGAGCCAGGTTCTGGCGA .....................................................(((((..............(((((..(((((((......)))))))..........(((....))).....)))))...((((((((........((....)).((((((((.......)))))))).))))))))...)))))..................................................... ..................................................51...................................................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTATCCTTTTTATTTGGCCATCGTCCTCAGGTGACTCATCCCAGTTACCACT.................................................................................................................................................... | 52 | 1 | 189.00 | 189.00 | 189.00 | - | - | - | - | - | - |
| .......................................................................................................................................................................TAGGTGAGAGGTTTTAGACACTGTTGGG....................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - |
| ...............................................................................................................................................................................................TGGGGAATTAATAGACTTCAAACATTCGT.............................. | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - |
| .......................TGTTCAATAGCACCCAGAGCCTCCTCT........................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - |
| TGGCTTCCTCTCAGGAGTCTCCATGTTCAATAGCACCCAGAGCCTCCTCTGTATCCTTTTTATTTGGCCATCGTCCTCAGGTGACTCATCCCAGTTACCACTCTTTCCACCTCTGTAGGCCTTTGGGGCACAAGTGTCTGCTCTACATGGTAAACCGAATCTCTCTCTAGGTGAGAGGTTTTAGACACTGTTGGGGAATTAATAGACTTCAAACATTCGTTAAATCCGGATGTGAGCCAGGTTCTGGCGA .....................................................(((((..............(((((..(((((((......)))))))..........(((....))).....)))))...((((((((........((....)).((((((((.......)))))))).))))))))...)))))..................................................... ..................................................51...................................................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .GGCTTCCTCTCAGGAGTCTCCATGTTCA............................................................................................................................................................................................................................. | 28 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | 1.00 |
| ..GCTTCCTCTCAGGAGTCTCCATGTTCAA............................................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - |
| .......................................................................................ATCCCAGTTACCACTCTTTCCACCT.......................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - |