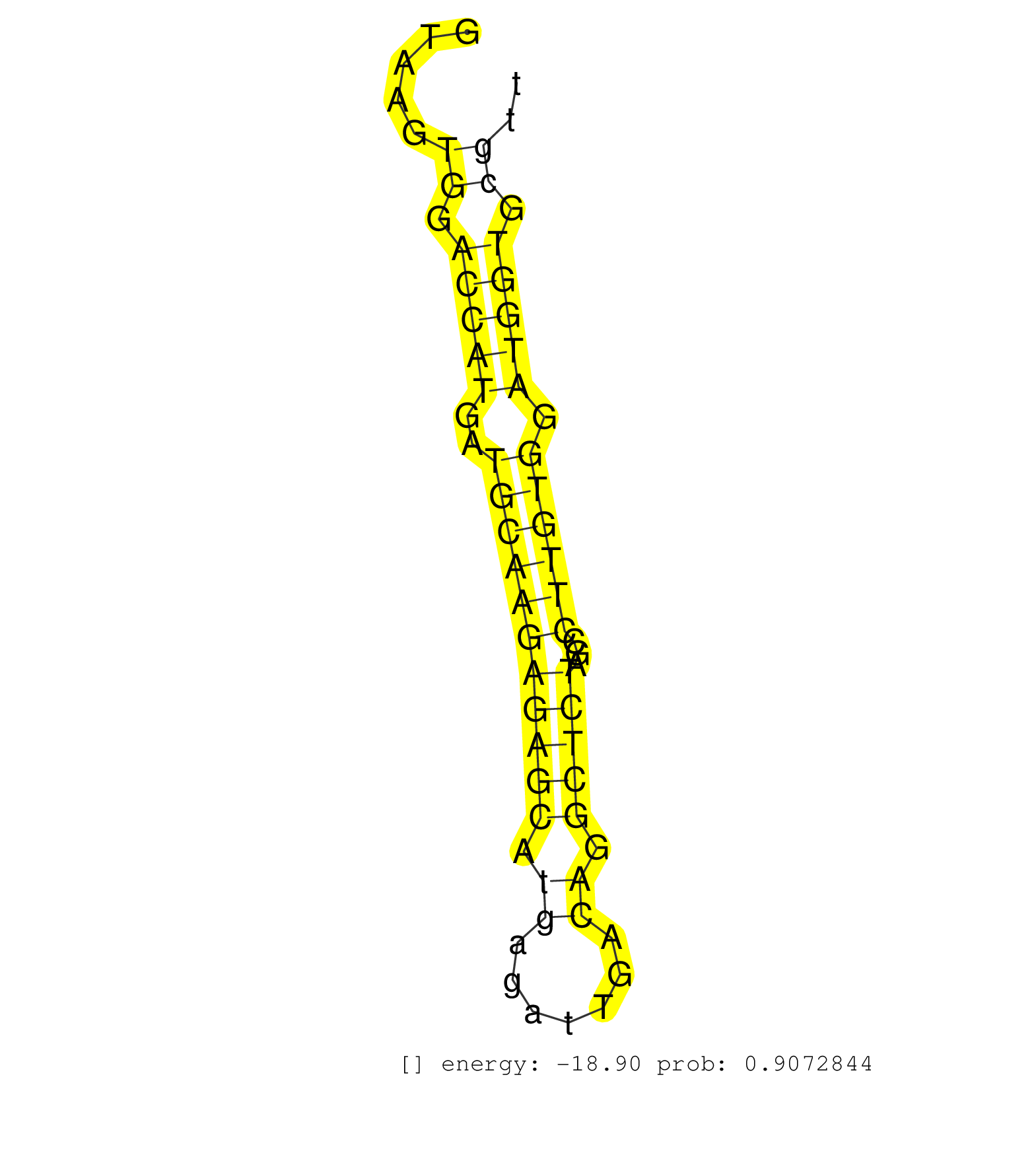

| Gene: Myh10 | ID: uc007jny.1_intron_18_0_chr11_68598859_f | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(2) PIWI.ip |
(2) PIWI.mut |
(17) TESTES |
| GATATCATCATCTTTTTCCAAGCTGTATGCAGAGGCTACCTCGCCCGAAAGTAAGTGGACCATGATGCAAGAGAGCATGAGATTGACAGGCTCTAGCCTTGTGGATGGTGCGTTCCCATGGGAGCAGCATCTGGATGAACAGATGCACATCCCAAACAACACACAGGCACCAGGAAAACACCGAGGGCCAAGAAATGAAACACAAGGCTTGGAGCTGTGTTTCAGCCGAGGTCCTGGGGAAGCCATGCCCCAGGGAGCTTTGCTGGAAGCCAGTCACACCCCTGAAGTGTGCTTTCCTTACTGTTCCTCAGGGCCTTTGCCAAGAAACAGCAACAACTAAGTGCCTTAAAGGTCTTGCAGC .......................................................((.(((((..(((((((((((.((.......)).)))))...)))))).))))).))......................................................................................................................................................................................................................................................... ..................................................51.............................................................114..................................................................................................................................................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | mjTestesWT3() Testes Data. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGTGGACCATGATGCAAGAGAGCA............................................................................................................................................................................................................................................................................................ | 27 | 1 | 8.00 | 8.00 | - | 1.00 | 2.00 | - | - | - | 1.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 |
| ..................................................GTAAGTGGACCATGATGCAAGAGAGCAT........................................................................................................................................................................................................................................................................................... | 28 | 1 | 7.00 | 7.00 | - | - | 1.00 | 1.00 | - | 2.00 | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - |
| ..................................................GTAAGTGGACCATGATGCAAGAGAG.............................................................................................................................................................................................................................................................................................. | 25 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................GTAAGTGGACCATGATGCAAGAGAGC............................................................................................................................................................................................................................................................................................. | 26 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................AAGTGGACCATGATGCAAGAGAGC............................................................................................................................................................................................................................................................................................. | 24 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTGGACCATGATG...................................................................................................................................................................................................................................................................................................... | 17 | 2 | 1.50 | 1.50 | - | - | - | - | - | - | - | - | - | - | - | 1.50 | - | - | - | - | - |
| ...................................................................................TGACAGGCTCTAGCCTTGTGGATGGTG........................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................TAAGTGGACCATGATGCAAGAGAGCATG.......................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................GTGGATGGTGCGTTtctt................................................................................................................................................................................................................................................... | 18 | tctt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...............TTCCAAGCTGTATGCAGAGGCTACCTCGC............................................................................................................................................................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............TTTCCAAGCTGTATGCAGAGGCTACCTC............................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAAGTGGACCATGATGCAAGAGAGCAT........................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................AGGCTACCTCGCCCGAAAGg..................................................................................................................................................................................................................................................................................................................... | 20 | g | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................AGTGGACCATGATGCAAGAGAGCATGt......................................................................................................................................................................................................................................................................................... | 27 | t | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TGTATGCAGAGGCTACCTC............................................................................................................................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| GATATCATCATCTTTTTCCAAGCTGTATGCAGAGGCTACCTCGCCCGAAAGTAAGTGGACCATGATGCAAGAGAGCATGAGATTGACAGGCTCTAGCCTTGTGGATGGTGCGTTCCCATGGGAGCAGCATCTGGATGAACAGATGCACATCCCAAACAACACACAGGCACCAGGAAAACACCGAGGGCCAAGAAATGAAACACAAGGCTTGGAGCTGTGTTTCAGCCGAGGTCCTGGGGAAGCCATGCCCCAGGGAGCTTTGCTGGAAGCCAGTCACACCCCTGAAGTGTGCTTTCCTTACTGTTCCTCAGGGCCTTTGCCAAGAAACAGCAACAACTAAGTGCCTTAAAGGTCTTGCAGC .......................................................((.(((((..(((((((((((.((.......)).)))))...)))))).))))).))......................................................................................................................................................................................................................................................... ..................................................51.............................................................114..................................................................................................................................................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | mjTestesWT3() Testes Data. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............TTCCAAGCTGTATGCAGAGGCTACCT................................................................................................................................................................................................................................................................................................................................ | 26 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................CCCATGGGAGCAGCAgggt........................................................................................................................................................................................................................................ | 19 | gggt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...................................................................................................................................................CATCCCAAACAACACACAGGCACCAGGA.......................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |