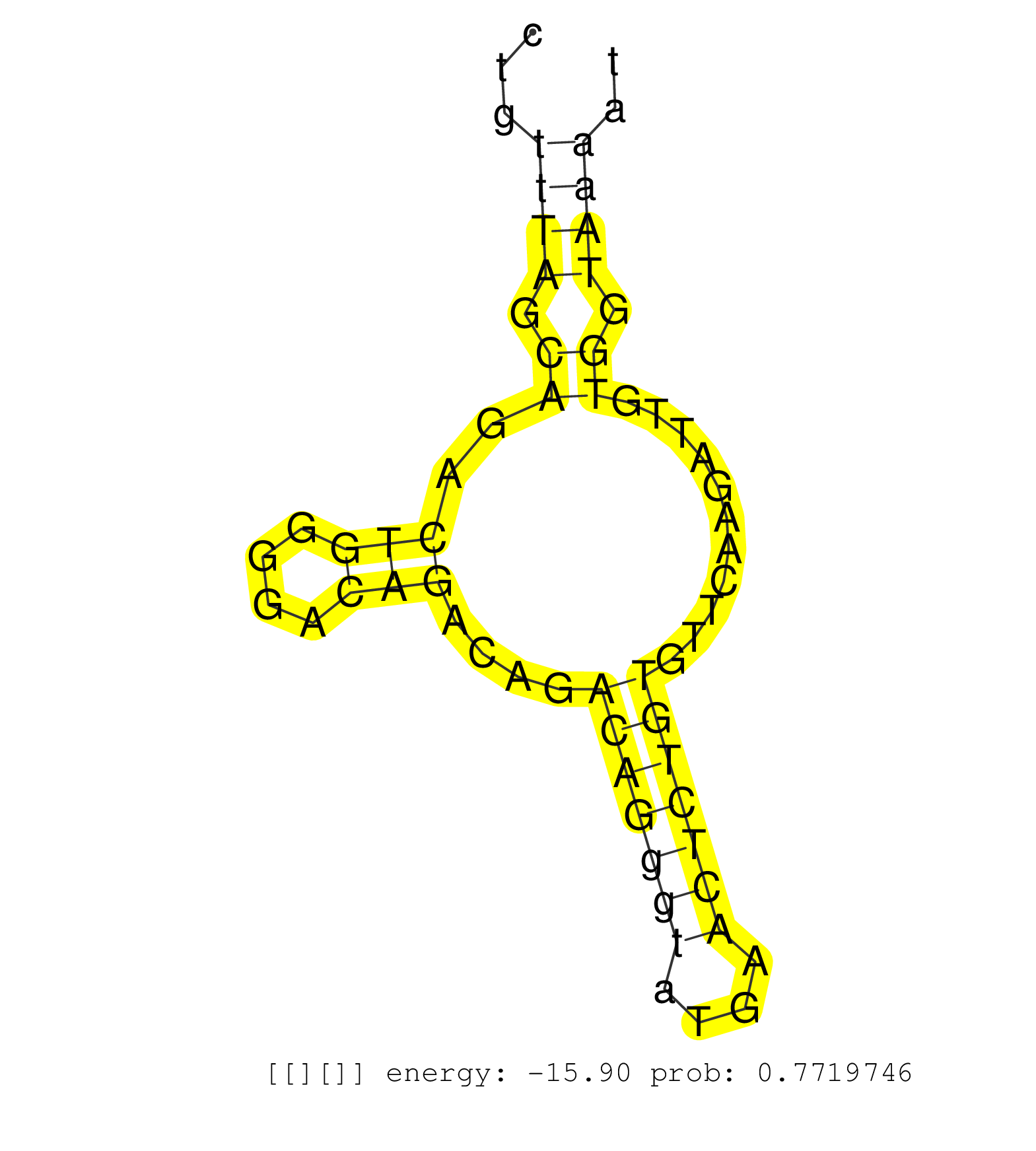

| Gene: Epn2 | ID: uc007jhx.1_intron_9_0_chr11_61379397_r.3p | SPECIES: mm9 |
 |
 |
|
(2) PIWI.ip |
(1) PIWI.mut |
(12) TESTES |
| AGGGCCATAACTTTGATATCCTCATGATGAAAAATATCCAAGATAAAAGTATAGCTGACCTCTGTAATCATTTTAAGTGTCAAGAAGCCTTTAGAATGTCTGTTTAGCAGACTGGGGACAGACAGACAGGGTATGAACTCTGTGTTCAAGATTGTGGTAAAATCAGTGAGATCTGTCTTTGCTTCTCCCTGCTCTCTCAGTTATGGCATTCCTTGCTCCAGGATTTGGGACAGGACGCTGATTTTGCACA ......................................................................................................((((.((..(((....)))....(((((((....)))))))...........)).))))......................................................................................... ...................................................................................................100.............................................................163..................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | mjTestesWT2() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................TGAACTCTGTGTTCAAGATTGTGGTA........................................................................................... | 26 | 1 | 7.00 | 7.00 | 2.00 | 1.00 | - | 2.00 | - | - | 1.00 | - | - | - | - | 1.00 |
| ...................................................................................................................................TATGAACTCTGTGTTCAAGATTGTGGTA........................................................................................... | 28 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - |
| ........................................................................................................TAGCAGACTGGGGACAGACAGACAG......................................................................................................................... | 25 | 1 | 3.00 | 3.00 | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TGAACTCTGTGTTCAAGATTGTGGTAA.......................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - |
| .............................................................................................................................................................TAAAATCAGTGAGATCTGTCTTTGC.................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................................TAGCAGACTGGGGACAGACAGAC........................................................................................................................... | 23 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ........................................................................................................TAGCAGACTGGGGACAGACAGACAGG........................................................................................................................ | 26 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................TGGCATTCCTTGCTCCAGGATTTGGGAC................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................................................................................TGTTCAAGATTGTGGTAAAATCAGTGA................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................TAAGTGTCAAGAAGCCTTTAGAATGTCTGT................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ........................................................................................................TAGCAGACTGGGGACAGACAGACA.......................................................................................................................... | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TGTGTTCAAGATTGTGGTA........................................................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................TGCTCTCTCAGTTATGGCATTCCTTGC.................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...........................TGAAAAATATCCAAGATAAAAGTATAGC................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TAGAATGTCTGTTTAGCAGACTGGGGAC................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................TTGCTCCAGGATTTGGGACAGGACGC............ | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TAGAATGTCTGTTTAGCAGACTGGGGACA.................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................TAGCAGACTGGGGACAGACAGACAGa........................................................................................................................ | 26 | a | 1.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................AGCAGACTGGGGACAGACAGACAGGGT...................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TAGAATGTCTGTTTAGCAGACTGGGGA.................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TTAGCAGACTGGGGACAGACAGACAGGGTA..................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| AGGGCCATAACTTTGATATCCTCATGATGAAAAATATCCAAGATAAAAGTATAGCTGACCTCTGTAATCATTTTAAGTGTCAAGAAGCCTTTAGAATGTCTGTTTAGCAGACTGGGGACAGACAGACAGGGTATGAACTCTGTGTTCAAGATTGTGGTAAAATCAGTGAGATCTGTCTTTGCTTCTCCCTGCTCTCTCAGTTATGGCATTCCTTGCTCCAGGATTTGGGACAGGACGCTGATTTTGCACA ......................................................................................................((((.((..(((....)))....(((((((....)))))))...........)).))))......................................................................................... ...................................................................................................100.............................................................163..................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | mjTestesWT2() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................................TGCTCTCTCAGTTATcat.............................................. | 18 | cat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |